Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Genetically encoded phenyl azide photochemistry drives positive and negative functional modulation of a red fluorescent protein†‡
Samuel C.
Reddington§
a,
Sarunas
Driezis§
a,
Andrew M.
Hartley§
a,
Peter D.
Watson
a,
Pierre J.
Rizkallah
b and
D. Dafydd
Jones
*a
aSchool of Biosciences, Cardiff University, Cardiff, UK. E-mail: jonesdd@cardiff.ac.uk; Tel: +44 (0)2920 20874290
bSchool of Medicine, Cardiff University, Cardiff, UK
First published on 8th September 2015
Abstract
The photochemical properties of phenyl azide have been exploited to modulate the function of a red autofluorescent protein, mCherry. Using genetic code reprogramming, phenyl azide chemistry has been introduced at functionally strategic positions in mCherry leading to deactivation, activation or enhancement upon UV irradiation.
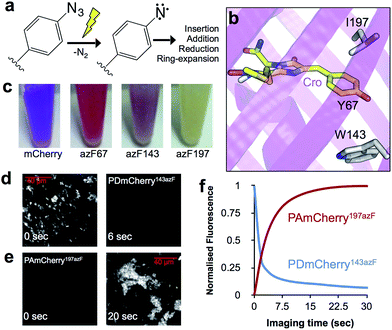
Optical control of biological processes whereby light is used to modulate inherent protein function is fast becoming an important tool in the biosciences as a means to precisely control protein activity with high temporal and spatial resolution not achievable through classical transcriptional control.1–3 Existing approaches to genetically encode protein photocontrol, known as optogenetics, generally utilise versions of natural light sensing proteins that detect and respond to light through the use of non-proteinaceous cofactors (e.g. retinal and flavins). The main drawback is the requirement for whole protein domains (e.g. LOV domains4 or membrane bound opsins2) to encode the light-responsive properties. An alternative approach is to use photoreactive chemical groups that can be programmed directly into a protein through the use of non-canonical amino acids (ncAAs).5 One such ncAA, p-azido-phenylalanine (azF; Fig. 1a), is particularly useful as it has a relatively small side-chain compared to other ncAAs (N3 at the para position instead of OH in the natural amino acid tyrosine) and can easily be photoconverted to a highly reactive nitrene radical (Fig. 1a).6,7 Subsequent products of the various nitrene reaction pathways can be used to alter protein structure and thus control function.8
Autofluorescent proteins are unique in nature in that they absorb and emit light in the visible range without the requirement of cofactors.9,10 Contiguous amino acids within the core of the protein covalently rearrange in the presence of O2 to form an extended conjugated π system that acts as a chromophore. Photoresponsive versions of autofluorescent proteins are critical to modern super-resolution microscopy.11 The two main classes of autofluorescent proteins are the green10 (primarily derived from the classical Aequorea victoria GFP) and red9 (primarily derived from the oligomeric Discosoma sp. DsRed) versions. While the two protein classes share a common β-can structure, their sequence identities are low with distinct chromophore structures, maturation and environment; all these factors contribute towards the significant differences in fluorescence between these autofluorescent proteins. Thus, useful mutations cannot always be simply transferred to the other. Red autofluorescent proteins are becoming the preferred option in cell imaging due to their lower excitation wavelength energy and reduced inherent cellular fluorescence background. While previous efforts have highlighted how ncAAs,12 including azF,13,14 can influence the properties of GFPs, little is known about their potential impact and use with respect to red versions. Here we address this using the widely used DsRed variant mCherry15 and show by replacing single pre-selected residues with azF how function can be either positively or negatively regulated by UV irradiation, both in vitro and in vivo.
Based on analysis of the mCherry structure [PDB 2H5Q16], residues in and around the chromophore were selected for replacement with azF (Fig. 1b and S1‡). Of the twelve different mutants generated, ten were produced as soluble protein, suggesting incorporation of azF within the protein core did not impact protein folding (Table S1‡). All but two of the soluble variants had a measureable fluorescent signal before and/or after UV irradiation, suggesting that azF is functionally tolerated within the core of the protein. On UV irradiation, fluorescence changes varied depending on the placement of azF and included decreases, increases and/or shifts in λEX and λEM (Table S1‡). While mCherry itself has a degree of sensitivity to UV light (Table S1 and Fig. S2‡), most of the photoresponsive azF mutants displayed a significantly greater magnitude of change. Three variants (Y67azF, W143azF and I197azF; Fig. 1b) exhibited the largest fold change in fluorescence and were selected for further characterisation.
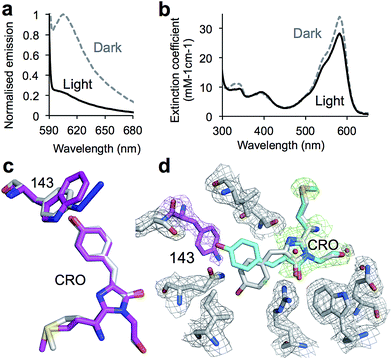
Replacement of W143 with azF (termed PDmCherry143azF, where PD refers to photodeactivation) generated a highly fluorescent protein in its dark state (pre-irradiation) that was responsive to UV light. Prior to irradiation, widefield microscopy of live E. coli cells indicated that PDmCherry143azF was relatively stable to photobleaching with a t1/2 of 76 s at 555 nm under typical imaging conditions for mCherry. The spectral peaks were slightly blue shifted (2–5 nm) compared to mCherry and it had a 20% lower quantum yield (Φ) and ∼50% lower molar extinction coefficient (ε) (Table 1). PDmCherry143azF was very sensitive to UV light (Fig. 1d), with live cell imaging revealing irradiation at 350 nm rapidly decreased observed fluorescence beyond the level inherent to mCherry (Fig. S2‡) with a t1/2 of 10 s. Detailed in vitro analysis confirmed these findings with PDmCherry143azF converting to a form with low fluorescence (Fig. 2a and Table 1). The absorbance spectrum indicated that photolysed protein still retained an entity that acts as a chromophore (λmax 582 nm), albeit with a slightly lower extinction coefficient (Fig. 2b).
| Variant | λ max (nm) | λ EX (nm) | λ EM (nm) | State | ε (M−1 cm−1) | Φ | Brightness M−1 cm−1 | Fold changea |
|---|---|---|---|---|---|---|---|---|
| a Fold change in fluorescence of variants was calculated from in vitro fluorescence spectra before and after photolysis. b Irradiated using a 5 W handheld lamp. c ND = not determined. The quantum yield of PAmCherryI197azF before photolysis was too low to be determined as protein absorbance (ε) was too low. d From N. C. Shaner, et al., Nat. Biotechnol., 2004, 22, 1567–1572. | ||||||||
| PDmCherryW143azF | 582 | 584 | 608 | Dark | 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.18 | 6120 | 5.1 |
| UVb | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
0.06 | 1548 | |||||
| PAmCherryI197azF | 584 | 587 | 610 | Dark | <1000 | NDc | — | ≥14.1 |
| UVb | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
0.05 | 580 | |||||
| mCherryd | 587 | 587 | 610 | — | 72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0.22 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 840 840 |
— |
To investigate the potential mechanism of action, the crystal structures of PDmCherry143azF resolved to 1.7 Å (dark; PDB code 4ZIN) and 2.0 Å (light; PDB code 4ZIO) were determined (see ESI for details‡). The structure of the dark state revealed that the variant has an essentially identical structure to mCherry with both the chromophore and residues at 143 occupying similar positions and planes (Fig. 2c). The packing around the chromophore was also very similar (Fig. S3‡). Thus, inserting a phenyl azide moiety into the core of the protein did not significantly disrupt structure. However, the presence of the azide group with its inherent resonance structures close to the chromophore may be influencing the latter's electronic properties.
Residue 143 lies close to the chromophore in mCherry (and the W143azF variant; Fig. 2c), occupying an equivalent position to Y/F145 in GFP, but both the backbone and the side chain placements differ significantly between the two (Fig. S4‡). Replacement of F145 in superfolder GFP (sfGFP17) with azF also led to loss of fluorescence on irradiation14 but by a different mechanism; the phenyl nitrene forms a crosslink to the chromophore so restricting chromophore mobility and reducing the extended conjugated double bond system. In contrast, the structure of irradiated PDmCherry143azF strongly suggests increased chromophore mobility is causing loss of fluorescence (Fig. 2d) with little change to the chromophore structure (Fig. 2b). The electron density for the irradiated form was generally poor over the chromophore region and residue 143, with no electron density observed for the p-hydroxybenzylidene chromophore moiety (Fig. 2d). The most likely explanation is the absence of a single defined conformation of the chromophore in the crystal, which suggests conformational flux of the chromophore upon UV irradiation of PDmCherry143azF. Isomerism and bond angle changes involving the chromophore are known to influence fluorescence.11 Indeed, the hydroxybenzylidene can be modeled to both the cis and trans conformation without steric problems or overlap with observed electron density (Fig. 2d). The limited electron density observed for residue 143 suggests a single atom protrusion at the para position, which we have tentatively assigned as the phenyl amine product of azF photolysis (Fig. 2d). The side chain of residue 143 points away from the chromophore, with the likely position of the nitrene radical being too far away (5.5–6 Å) to form a crosslink with the chromophore (Fig. 2d). The similar absorbance spectra for both dark and light forms of PDmCherry143azF suggest that the chemical structure is not significantly perturbed on irradiation. Otherwise, the structures of the dark and light states are very similar (Fig. S3‡) highlighting the crucial role relatively small structural changes instigated by azF photochemistry have on modulating fluorescent protein function and local dynamics.
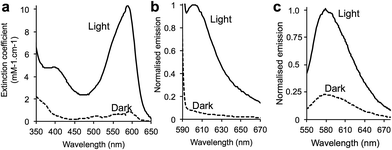
Replacement of I197 with azF resulted in the production of a non-fluorescent protein (termed PAmCherry197azF, where PA refers to photoactivation) with little inherent colour (Fig. 1c), cellular fluorescence (Fig. 1d) or absorbance (Fig. 3a). Neither the Φ nor ε for the non-irradiated form could be accurately calculated, suggesting that chromophore maturation had been impeded on production of the protein in the dark. Live cell imaging by widefield microscopy revealed that PAmCherry197azF could be rapidly activated by UV irradiation, with an observed t1/2 at 350 nm of 10 s (Fig. 1d). UV irradiated PAmCherry197azF showed higher apparent photostability than native mCherry, a key characteristic of photoactivatable fluorescent proteins (compare Fig. 1f and S2b‡); no apparent decrease in cellular fluorescence was observed after 60 s of imaging. More detailed in vitro studies demonstrated that on irradiation, fluorescence increased ∼14 fold in intensity with a final λEX and λEM similar to mCherry (Fig. 3a and Table 1). While irradiation resulted in functional activation, presumably through inducing chromophore maturation, the irradiated protein was less bright than native mCherry (Table 1) but could still imaged by microscopy (Fig. 1e).
I197 lies just above the plane of the chromophore in mCherry at an equivalent structural position to T203 in GFP (Fig. S5‡). Together with numerous additional mutations, substitution of I197 (to arginine) has previously contributed towards the generation of useful photoactivatable mCherry variants.18 In contrast to the observations here for PAmCherryI197azF, replacement of T203 in sfGFP with azF did not appear to significantly affect chromophore maturation but did red shift excitation and emission in the dark.13 The absence here of a red shift in spectral properties suggests that PAmCherry197azF does not display aromatic stacking between residue 197 and the chromophore. Also, photolysis of sfGFP T203azF results in a major reduction in extinction coefficient (∼5 fold)13 whereas substantial increase is observed for PAmCherryI197azF (Table 1). Thus, in this case the influence of azF and its photochemistry on function is not simply transferred from sfGFP to mCherry highlighting the role played by the local chemical environment.
I197 together with K70 are known to have an important role in chromophore maturation in mCherry,19 especially concerning the Y67 Cα–Cβ oxidation step, with K70 acting as a general base for proton abstraction.20 Given the proximity of K70 and I197 in space (Fig. S5‡), replacement of residue 197 is likely to disrupt the positioning of K70, so preventing it from acting as a general base. It is now thought that chromophore maturation of DsRed derived proteins (such as mCherry) proceed via a blue emitting intermediate, equivalent to the conjugated imidazolinone and acylimine groups.19,20 Loss of the potential Cα–Cβ oxidation step does not explain the apparent lack of this intermediate form in the dark state. This suggests that prior to irradiation, chromophore maturation progresses to, at most, the formation of the initial cyclised non-oxidised form (through the linking of residues M66 and G68). One hypothesis is that the phenyl azide group alters the positions of key residues such as K70 involved in chromophore maturation at both the Y67 Cα–Cβ and the M66 N–Cα oxidation steps required for a completed conjugation system. It could be possible that loss of molecular N2 (and associated local structural rearrangements) together with the formation of the nitrene radical itself and/or a final reaction product (e.g. phenyl amine) is involved in the maturation pathway, substituting for or modulating the role played by residues such as K70 in mCherry. Unfortunately, we were unable to crystallise this variant. It is important to note that it is the azide moiety that is likely to play the key role in controlling maturation in PAmCherry197azF as mutation to tyrosine (I197Y mutant) does not affect mCherry maturation fluorescence (Fig. S6‡).
The mCherry Y67azF variant (equivalent to Y66 in GFP), termed PEmCherry67azF (where PE refers to photoenhancement), has the tyrosine within the chromophore replaced. The transmissive properties of PEmCherry67azF were very different to mCherry with cell lysates turning from a purple to red colour (Fig. 1c), with replacement of the phenol group with phenyl azide in the chromophore the likely cause (vide infra). PEmCherry67azF was produced as a weakly fluorescent protein with the emission peak blue shifted by ∼50 nm compared to mCherry (Table S1‡), which is in keeping with previous observations that the electron rich azido group can act as a fluorescence quencher.8,21 On UV irradiation with low power UV light, a 3.4 fold increase in fluorescence was observed, with the blue shift in λEX and λEM compared to mCherry retained (Fig. 3b and Table S1‡). Previous work with GFP suggests that this is caused by the conversion of the phenyl azide to a phenyl amine.14 Replacing azF at residue 67 with p-amino-L-phenylalanine to mimic the potential photochemical endpoint produced a protein with similar maximal excitation and emission wavelengths to that of the irradiated PEmCherry67azF (Fig. S7‡), supporting the idea of conversion to the phenyl amine. The exchange of the phenol, or more accurately phenolate to phenyl amine changes the electron donation strength of the aromatic component and thus the resonant structures of the chromophore. This in turn could cause the observed blue shift in fluorescence.
The genetic incorporation of phenyl azide chemistry into proteins is a potentially general tool for controlling protein activity through the use of light without the need for bulky protein domains and additional cofactors. As demonstrated here with the widely used autofluorescent protein mCherry, the position within the protein's molecular structure is critical to how the potential photochemical pathway taken will ultimately impact function. Through incorporation of a single ncAA at three disparate positions in the protein core, we have generated PD, PA and PE mCherry variants, that could be developed into useful tools for microscopy. FRAP (fluorescence recovery after photobleaching) microscopy22 is normally hindered by the photostability of autofluorescent proteins meaning long and relatively intense illumination are required to deactivate the protein. This results in the loss of temporal resolution. PDmCherry143azF is relatively photostable at imaging wavelengths but can be rapidly deactivated at low light energy making it potentially an excellent probe for furthering FRAP microscopy. PALM (photoactivated localisation microscopy) requires photoactivatable fluorescent proteins with high levels of photostability,23,24 a characteristic we have successfully introduced into PAmCherry197azF. While photocontrol of autofluorescent proteins (including mCherry18) require multiple canonical mutations to achieve useful light-sensitive properties, we have shown here that incorporation of a single light sensitive ncAA can achieve a similar endpoint and sample different photochemical effects. Improved approaches for efficient and higher yielding ncAAs containing proteins in different cells and organisms25 combined with the inherent chemical genetic element (ncAA-dependent protein production) will broaden the potential use with regards to autofluorescent proteins and proteins in general.
Notes and references
- K. Deisseroth, Nat. Methods, 2011, 8, 26–29 CrossRef CAS PubMed.
- A. Gautier, C. Gauron, M. Volovitch, D. Bensimon, L. Jullien and S. Vriz, Nat. Chem. Biol., 2014, 10, 533–541 CrossRef CAS PubMed.
- J. E. Toettcher, C. A. Voigt, O. D. Weiner and W. A. Lim, Nat. Methods, 2011, 8, 35–38 CrossRef CAS PubMed.
- A. Moglich and K. Moffat, Photochem. Photobiol. Sci., 2010, 9, 1286–1300 Search PubMed.
- A. Gautier, A. Deiters and J. W. Chin, J. Am. Chem. Soc., 2011, 133, 2124–2127 CrossRef CAS PubMed.
- J. Chin, S. Santoro, A. Martin, D. King, L. Wang and P. Schultz, J. Am. Chem. Soc., 2002, 124, 9026–9027 CrossRef CAS PubMed.
- S. Reddington, P. Watson, P. Rizkallah, E. Tippmann and D. D. Jones, Biochem. Soc. Trans., 2013, 41, 1177–1182 CrossRef CAS PubMed.
- J. L. Morris, S. C. Reddington, D. M. Murphy, D. D. Jones, J. A. Platts and E. M. Tippmann, Org. Lett., 2013, 15, 728–731 CrossRef CAS PubMed.
- A. Miyawaki, D. M. Shcherbakova and V. V. Verkhusha, Curr. Opin. Struct. Biol., 2012, 22, 679–688 CrossRef CAS PubMed.
- R. Y. Tsien, Annu. Rev. Biochem., 1998, 67, 509–544 CrossRef CAS PubMed.
- D. M. Shcherbakova and V. V. Verkhusha, Curr. Opin. Chem. Biol., 2014, 20, 60–68 CrossRef CAS PubMed.
- W. Niu and J. Guo, Mol. BioSyst., 2013, 9, 2961–2970 RSC.
- S. C. Reddington, A. J. Baldwin, R. Thompson, A. Brancale, E. M. Tippmann and D. D. Jones, Chem. Sci., 2015, 6, 1159–1166 RSC.
- S. C. Reddington, P. J. Rizkallah, P. D. Watson, R. Pearson, E. M. Tippmann and D. D. Jones, Angew. Chem., Int. Ed., 2013, 52, 5974–5977 CrossRef CAS PubMed.
- N. C. Shaner, R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer and R. Y. Tsien, Nat. Biotechnol., 2004, 22, 1567–1572 CrossRef CAS PubMed.
- X. Shu, N. C. Shaner, C. A. Yarbrough, R. Y. Tsien and S. J. Remington, Biochemistry, 2006, 45, 9639–9647 CrossRef CAS PubMed.
- J. D. Pedelacq, S. Cabantous, T. Tran, T. C. Terwilliger and G. S. Waldo, Nat. Biotechnol., 2006, 24, 79–88 CrossRef CAS PubMed.
- F. V. Subach, G. H. Patterson, S. Manley, J. M. Gillette, J. Lippincott-Schwartz and V. V. Verkhusha, Nat. Methods, 2009, 6, 153–159 CrossRef CAS PubMed.
- F. V. Subach, V. N. Malashkevich, W. D. Zencheck, H. Xiao, G. S. Filonov, S. C. Almo and V. V. Verkhusha, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 21097–21102 CrossRef CAS PubMed.
- F. V. Subach and V. V. Verkhusha, Chem. Rev., 2012, 112, 4308–4327 CrossRef CAS PubMed.
- K. Sivakumar, F. Xie, B. M. Cash, S. Long, H. N. Barnhill and Q. Wang, Org. Lett., 2004, 6, 4603–4606 CrossRef CAS PubMed.
- A. Miyawaki, Nat. Biotechnol., 2004, 22, 1374–1376 CrossRef CAS PubMed.
- M. Fernandez-Suarez and A. Y. Ting, Nat. Rev. Mol. Cell Biol., 2008, 9, 929–943 CrossRef CAS PubMed.
- J. Lippincott-Schwartz and G. H. Patterson, Trends Cell Biol., 2009, 19, 555–565 CrossRef CAS PubMed.
- J. W. Chin, Annu. Rev. Biochem., 2014, 83, 379–408 CrossRef CAS PubMed.
Footnotes |
| † The PDB accession codes for dark and irradiated PDmCherry143azF are 4ZIN and 4ZIO. We would like to thank the staff at the Diamond Light Source for the supply of facilities and beam time, especially Beamline I03 and I04 staff. We thank BBSRC (BB/H003746/1 and BB/M000249/1), EPSRC (EP/J015318/1) and Cardiff SynBio Initiative/SynBioCite for supporting this work. |
| ‡ Electronic supplementary information (ESI) available: Detailed methods, supporting Fig. S1–S7, supporting Tables S1–S3. See DOI: 10.1039/c5ra13552d |
| § All these authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |