Causal inference methods to assist in mechanistic interpretation of classification nano-SAR models†
Natalia Sizochenkoab,
Bakhtiyor Rasulevbc,
Agnieszka Gajewicza,
Elena Mokshynad,
Victor E. Kuz'mind,
Jerzy Leszczynskib and
Tomasz Puzyn*a
aLaboratory of Environmental Chemometrics, Faculty of Chemistry, University of Gdansk, Wita Stwosza 63, 80-308, Gdansk, Poland. E-mail: t.puzyn@qsar.eu.org
bInterdisciplinary Center for Nanotoxicity, Department of Chemistry, Jackson State University, 1400 J. R. Lynch Street, P. O. Box 17910, Jackson, MS 39217, USA
cCenter for Computationally Assisted Science and Technology (CCAST), North Dakota State University, 1805 NDSU Research Park Dr, PO Box 6050, Fargo, ND 58108, USA
dA.V. Bogatsky Physical–Chemical Institute National Academy of Sciences of Ukraine, 65080, Odessa, Ukraine
First published on 8th September 2015
Abstract
Knowledge about the toxicity of nanomaterials and factors responsible for such phenomena are important tasks necessary for efficient human health protection and safety risk estimation associated with nanotechnology. In this study, the causation inference method within structure-activity relationship modeling for nanomaterials was introduced to elucidate the underlying structure of the nanotoxicity data. As case studies, the structure-activity relationships for toxicity of metal oxide nanoparticles (nano-SARs) towards BEAS-2B and RAW 264.7 cell lines were established. To describe the nanoparticles, the simple ionic, fragmental and “liquid drop model” based descriptors that represent the nanoparticles' structure and characteristics were applied. The developed classification nano-SAR models were validated to confirm reliability of predicting toxicity for all studied metal oxide nanoparticles. Developed models suggest different mechanisms of nanotoxicity for the two types of cells.
Introduction
For the last decade researchers have dedicated considerable efforts towards synthesis and experimental analysis of various nanomaterials and development of species characterized by different, desired properties.1,2 As is well known, peculiar physical and chemical properties of nanoparticles occur as a manifestation of surface and quantum size effects.3 For many complex chemical entities, experimental testing of nanoparticles is costly and time-consuming.4 This fact limits the development and further testing of new nanoparticles with better properties. One of the very fast alternative solutions is the in vitro testing using different types of cells and cell lines.5 However, the problem lies in insufficiency and inconsistency of experimental toxicity data for nanoparticles.6 Moreover, this problem is amplified by differences between criteria and protocols developed for various cell lines, differences in methods of expressing concentration of nanoparticles and the procedures for the estimation of toxicological effects. For some nanoparticles data is available in reports performed by different laboratories. As a consequence, different results in different reports are obtained for the same composition, where the diversity could be due to the methods for handling nanoparticles and preparing dispersions, variation in size distribution and dispersion stability, and different exposure conditions. Considering the important role of different factors in the diversity of results, it is essential to characterize nanoparticles in the relevant medium and to apply appropriate treatment conditions. In this connection, intra-laboratory reproducibility should at least be evaluated in order to obtain robust results.7Simultaneously, there is an increasing interest towards in silico prediction of activity and toxicity.8–18 The (Quantitative) Structure-Activity Relationship ([Q]SAR) modeling plays currently an important role as an efficient tool for various properties prediction. However, the main purpose of this approach is not only development of (Q)SARs that have predictive power, but also obtaining models maintaining the ability of mechanistic interpretation.9,13,16
The well-known phrase says “Correlation is not causation”, and traditional approaches towards the interpretation do not show existing “cause-effect” relationships. Causal molecular interactions can be discovered using randomized experiments; however such experiments are often costly, infeasible, or limited by biological ethical issues. Algorithms that infer causal interactions have only recently been applied to genomics data,19 but never before proposed within the framework of (Q)SAR studies.
The aim of current study is to demonstrate usefulness of methods of causal discovery to elucidate the underlying structure of the nanotoxicity data and retrieve additional, more robust interpretation for the developed (Q)SAR models. To make this, we have developed nano-SAR models for toxicity of metal oxides nanoparticles towards BEAS-2B cells and RAW 264.7 cells using simple geometric, fragmental and physical descriptors and applied the causal inference methods to provide mechanistic interpretation of results.
Theory of causal inference methods
Causal inference models can be approximately described as in silico way to match the notion of randomized controlled experiment, which is the standard for evaluating causation scientifically.20,21 However, as indicated above, real-life randomized experiments are costly, time-consuming and can be unethical. Let us assume that in a hypothetical experiment, there is a certain distribution of a variable X. It is implied that X is a cause of Y (and Y is a dependent of X) and denote this by X → Y if the probability distribution of Y changes for some experimental manipulation of X.During the last two decades many algorithms that infer causal interactions from observational data have been developed.22–27 An excellent review discussing the details can be recommended.22
Causal inference methods are based on several fundamental mathematical concepts, which include conditional probability and its joint distribution, directed and undirected graphs. As these are fundamental and wide concepts, format of this article does not allow describing them in details, but some basics are provided below.
A conditional probability measures the probability of an event given that (by assumption, presumption, assertion or evidence) another event has occurred. If the events are denoted as A and B, respectively, this is said to be “the probability of A given B”. In statistical inference, the conditional probability is an update of the probability of an event based on new information. It is commonly denoted by P(A|B), or sometimes PB(A). Given two jointly distributed random variables X and Y, the conditional probability distribution of Y given X is the probability distribution of Y when X is known to be a particular value.
The purpose of the tested causal orientation methods is to separate the cause from the effect of given data for just two variables X and Y that have a causal relation (i.e., in the underlying data generative distribution, either X → Y or X ← Y). Most of these techniques are based on the idea that the factorization of the joint probability distribution P(cause, effect) into P(cause)P(effect|cause) yields a simpler representation than the factorization into P(effect)P(cause|effect). One can furthermore show that, if the marginal probability distribution of the cause – P(cause) is independent of the causal mechanism P(effect|cause), then the factorization P(cause)P(effect|cause) has lower complexity than the factorization P(effect)P(cause|effect). Given two causally related variables X and Y, estimating the complexity of the two different factorizations of P(X, Y) or determining independence between marginal and conditional distributions can thus provide the basis for causal orientation techniques.
In practice, however, it is difficult to directly test independence between P(X) and P(Y|X) or estimate (or even define a measure of) their complexity; hence the methods typically use simplifying assumptions or rely on approximate formulations. Some causal orientation methods output two scores indicating likelihood of the forward causal model (X → Y) and the backward one (X ← Y). Other methods output two p-values indicating significance of the forward and backward causal models.
Well-known Peter Spirtes and Clark Glymour (PC) algorithm24 is based on conditional independence tests. To apply it, sufficient statistics should be calculated and a conditional independence test function specified. PC starts with a complete undirected graph (Fig. 1).
Then, a series of conditional independence tests is done and edges are deleted. The result is a skeleton, in which every edge is still undirected. In the next step orientation of edges is found by repeatedly applying rules (i.e. one can deduce that one of the two possible directions of the edge is invalid because it introduces a directed cycle). Hypothetic directed graph obtained based on the above approach is presented in Fig. 2.
As we can see, the map here represents a graph consisting of nodes and directed edges (← or →). Fig. 3 depicts possible relations. First case refers to A as a cause of B, second to B as a cause of A, in the third case variables A and B have the common cause C, but are independent, the fourth case describes two independent variables, and in the fifth case nothing can be implied about the direction of causal relationship.
Unfortunately more detailed description of the methods used is beyond the scope of this paper. The relevant articles and source code are available on the website of the Max-Planck-Institute for Intelligent Systems Tübingen.
Case study: toxicity of metal oxide nanoparticles
Materials and methods
• Simplex Representations of Molecular Structure (SiRMS)-based descriptors;29
• Metal-ligand binding descriptors;30,31
• “Liquid drop” model (LDM) derived descriptors;16,32
• Integral (constitutional) descriptors for each molecule, such as molecular weight, mass density and aligned electronegativity of oxide.
Simplex representation of molecular structure (SiRMS). In the current study we utilized a 2D level of Simplex Representation of Molecular Structure (SiRMS)29 theory to generate two-, tri- and tetra-atomic molecular fragments (simplexes). Atomic electronegativity, the Lennard-Jones potential and partial charges were expressed within intervals. For readers who interested in calculating SiRMS-based descriptors the in-house software can be provided by request.
Metal-ligand binding (MLB) characteristics. To get a complete picture describing the toxic action, descriptors related to ionic characteristics were utilized. Metal-ligand binding (MLB) theory assumes that binding of metals to soft ligands at biomolecules plays an important role in exhibition of toxicity.30,31 In the current study, two ion characteristics were used: covalent index (CI) and cation polarizing power (CPP).
“Liquid drop” model (LDM). To describe the geometric and volume features of studied nanoparticles, the physical model of “liquid drop” was used.32,34 Within LDM the minimum radius of the interactions between elementary particles is described by so-called Wigner-Seitz radius.32,34 Shape of nanoparticle is represented as a spherical drop. Based on geometric nature of LDM, transformations allowed calculating the number of molecules in the nanocluster, the volume of the nanoparticle, aggregation parameter, the nanoparticle's surface area, the number of surface molecules and the surface-area-to-volume ratio. More details on these transformations are presented in our recent paper.16
Several calculated descriptors in current study were the same as in Liu et al. paper.33 All the formulas and full list of utilized descriptors are presented in ESI 1 and 2.†
RF is an ensemble classifier proposed by Breiman.36 It constructs a series of decision trees. Several developed decision trees are combined to consensus forest. Influence of each descriptor has the relative value on the basis of the average of the individual tree predictions.36
Initial datasets were splitted into training and test sets. Quality of developed models were analyzed both for training and test sets using several statistical measures:
 | (1) |
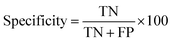 | (2) |
 | (3) |
Domain applicability (DA) was measured based on minimum-cost-tree of variable importance values in space of descriptors considering their relative importance.37
Correlation between target properties was evaluated via φ-coefficient:38
 | (4) |
Results and discussion
Model development
The information about correlation between two binary sequences of toxicity (for BEAS-2B and RAW264.7 cells) was summarized by defining the four dependent quantities (Table 1). Since φ-coefficient for investigated data of toxicity is equal 0.51, there is an average degree of association between two types of toxicity.| BEAS-2B | RAW 264.7 cells | |
|---|---|---|
| Value 1 | Value 0 | |
| Value 1 | 8 (a) | 4 (b) |
| Value 0 | 2 (c) | 10 (d) |
After calculating all the descriptors, variables having zero-variance, and highly cross-correlating variables (with the Pearson's pair correlation coefficient |r| > 0.9) were eliminated.
Nanoparticles were splitted into training and test set (18 and 6 compounds, respectively) in the following way - the splitting of the dataset to training and test sets fulfilled three conditions: (1) metal oxides from each activity group should be presented in both training and test sets; (2) metal oxides presented in the test set should cover all types of oxides (MeO, Me2O3, MeO2), similarly to the training set; (3) the list of oxides in each test set should be identical for both toxicity endpoints.
Then, QSAR tasks were processed using Random Forests regression (5 trees, 5 descriptors in each). In both cases (BEAS-2B and RAW 264.7) the number of true negative classifications (specificity) and for the training sample corresponds with the value of those in the original sample. Model for BEAS-2B also has absolute (100%) sensitivity and balanced accuracy. This means that model for BEAS-2B represents an ideal case where the model accurately determines the class with absolute probability (hundred true results of a hundred). In case of model for RAW264.7, there was one false prediction of toxicity (toxic Yb2O3 was predicted as non-toxic – see Table 2). Thus, sensitivity of training set for RAW 264.7 model was 88% and balanced accuracy was 94%.
| No. | Metal oxide NP | BEAS-2B (observed/predicted) | BEAS-2B (DA) | RAW 264.7 (observed/predicted) | RAW 264.7 (DA) |
|---|---|---|---|---|---|
| a Test set compounds are marked in bold. | |||||
| 1 | Al2O3 | 0/0 | + | 0/0 | + |
| 2 | CuO | 1/1 | + | 1/1 | + |
| 3 | CeO2 | 1/1 | + | 0/0 | + |
| 4 | Co3O4 | 1/1 | + | 1/1 | + |
| 5 | CoO | 1/1 | + | 1/1 | + |
| 6 | Cr2O3 | 1/1 | + | 1/1 | + |
| 7 | Fe2O3 | 0/0 | + | 0/0 | + |
| 8 | Fe3O4 | 1/1 | + | 0/0 | + |
| 9 | Gd2O3 | 0/0 | + | 0/0 | + |
| 10 | HfO2 | 0/0 | + | 1/1 | + |
| 11 | In2O3 | 0/0 | + | 0/0 | + |
| 12 | La2O3 | 0/1 | + | 0/0 | + |
| 13 | Mn2O3 | 1/1 | + | 1/1 | + |
| 14 | NiO | 1/1 | + | 1/1 | + |
| 15 | Ni2O3 | 1/1 | + | 1/1 | + |
| 16 | Sb2O3 | 0/0 | + | 0/0 | + |
| 17 | SiO2 | 1/1 | + | 0/0 | + |
| 18 | SnO2 | 0/0 | + | 0/0 | + |
| 19 | TiO2 | 0/0 | + | 0/0 | + |
| 20 | Y2O3 | 1/0 | + | 0/0 | + |
| 21 | Yb2O3 | 0/0 | + | 1/0 | + |
| 22 | ZnO | 1/1 | + | 1/1 | + |
| 23 | ZrO2 | 0/0 | + | 0/1 | + |
| 24 | WO3 | 0/0 | + | 0/0 | + |
The predictive ability of the obtained models was estimated on a test set. For test samples, the sensitivity (ratio of true positive classifications) was also 100% for both toxicity endpoints. However, specificity (the ratio of true negative classifications) in the case of RAW 264.7 was slightly higher (75%) than that in the case of BEAS-2B (66%).
In the case of theoretical analysis of toxicity such sensitivity values mean that developed models with a probability of 25% (100% values of initial data minus 75% of true negative classifications) and 33% (100% values of initial data minus 66% of true negative classifications) do positively predict the toxicity of new entities for BEAS-2B and RAW 264.7, respectively. Since theoretical analysis methods are mainly preclinical, we allow final models to assign toxic mark for some ambiguous compounds to prevent toxic compounds be marked as non-toxic. The balanced accuracy of test sets was 83% and 88% for BEAS-2B and RAW264.7 models, respectively.
In Table 2 the data about domain applicability, observed and predicted values are provided. More details about developed models are available in ESI 2.†
Model interpretation
Interpretation and analysis of the classification models allowed us to identify a number of structural parameters responsible for nanoparticles toxicity. Classification model for BEAS-2B includes 7 descriptors, whereas model for RAW 264.7 uses 9 descriptors (Table 3).| Group of descriptors | Type of descriptor | Model | |
|---|---|---|---|
| BEAS-2B | RAW 264.7 | ||
| a Numbers indicate relative influence of descriptors in Random Forest models. | |||
| Integral parameters | Mass density | 0.100 | 0.022 |
| Molecular weight | 0.022 | ||
| Aligned electronegativity | 0.011 | ||
| MLB characteristics | Covalent index | 0.083 | 0.044 |
| Cation polarizing power | 0.017 | ||
| LDM-based descriptors | Wigner-Seitz radius | 0.033 | |
| Surface area | 0.033 | ||
| Surface-area-to-volume ratio | 0.050 | 0.011 | |
| Aggregation parameter | 0.067 | ||
| SiRMS descriptors | Two-atomic descriptor of van-der-Waals interactions | 0.033 | |
| Tri-atomic descriptor of atomic charges | 0.05 | ||
| Tetra-atomic descriptor of atomic charges | 0.011 | ||
| Size in DMEM | 0.044 | ||
Difference in the structural parameters used in both models suggests that the mechanisms of the toxicity of nanoparticles towards both studied cell cultures are also different. Both models use mass density. This is fundamental property which is utilized in the LDM approach. Covalent index (CI) reflecting interactions of nanoparticles with protein-bound sulfhydryl's and depleting glutathione has been selected as an important variable in a model of toxicity to BEAS-2B cells. Cation polarization power (CPP) in both models reflects electrostatic interactions between nanoparticles and cells. Also this is in agreement with the impact of electronegativity descriptor in model for RAW 264.7.
We suppose that surface area and surface-area-to-volume ratio in the model indirectly describe the ability of nanocluster's surface molecules fraction to leach from the surface of nanoparticle. As it is known, surface molecules are more reactive and facilitate the massive oxidizing capabilities.39
The aggregation factor (BEAS-2B model) may indirectly describe the mechanism of penetration of nanoparticles into biological systems, which links damage of organelles, depending on the size of nanoparticles.35 In the same way, for RAW 264.7 the size in DMEM can reflect the mechanism of toxicity.
Causal structures
In the next step of the study, the causal structures were implemented to compare our results with previously published data (Fig. 4 and 5). To provide causal analyses, all available descriptors were used.Let's take a look at developed causal structures. In both cases, the electronegativity is mutually related to the point of zero zeta-potential (Fig. 4 and 5). In case of BEAS-2B cells, Ec has mutual relations with SiRMS charges descriptor, wherein Ec and PZZP have direct relationships with target toxicities (Fig. 4). For RAW 264.7 the triatomic SiRMS charges have mutual relationship with size of single nanoparticle (Fig. 5). Additionally, the size of a single nanoparticle is related to target toxicity.
There are no causal relations between target properties and MLB characteristics. Also not all descriptors included in the developed models are causally-dependent with target properties and with relation to each other.
In fact, there is no direct link between toxicity and any descriptor. It means there are only particular causal links and the developed models are the collection of the most important descriptors, which only represent the conditions for the emergence of particular cause of action.
Conclusions
In conclusion, it can be stated that the causal inference method presented in this paper can lead to many valuable observations and conclusions. The present study demonstrates that causal structures can efficiently be used in nano-SAR modeling as additional criteria for quality evaluation. Two classification nano-SAR models were validated in terms of causal inference. In addition we have shown that introducing causation methods into structure-activity relationship modeling for nanomaterials allows to elucidate underlying structure of the nanotoxicity data. A great advantage of presented method is the fact that it provides the mechanistic interpretation of obtained results. In other words, methods of causal discovery open new opportunities to provide useful information at the molecular level that could be used to reveal mechanisms of toxicity.For the model development we have utilized a computational modeling methodology to build computational classification models for quick predictions of the ranks of toxicity. By applying causal inference methods it was proved that the proposed descriptors and statistical approach provide the convenient and efficient tool for prediction of nano-sized metal oxides toxicity. The obtained results reveal some new aspects of the biological action.
We assume that the causal structures are very promising tool not only for modeling of biological activity, but also for a variety of other properties of nanoparticles which are due to the peculiarities of nanostructures. In addition, data visualization techniques in causal analyses are very helpful to understand special interactions between important descriptors in different models.
Acknowledgements
The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement #309837 (NanoPUZZLES project). This work was partially funded by the Foundation for the Polish Science within FOCUS program. The authors thank for the financial support the European Commission through the Marie Curie IRSES program, NanoBRIDGES project (FP7-PEOPLE-2011-IRSES, grant agreement #295128). N.S. and J.L. thank the National Science Foundation for support from the NSF CREST Interdisciplinary Nanotoxicity Center – grant #HRD-0833178. B.R. gratefully acknowledges support from the North Dakota State University Center for Computationally Assisted Science and Technology and the U.S. Department of Energy through Grant No. DESC0001717.References
- D. Vollath, Nanomaterials: an introduction to synthesis, properties and applications, John Wiley & Sons, 2013, pp. 1–12 Search PubMed.
- G. Cao, Nanostructures & Nanomaterials: Synthesis, Properties and Applications, World Scientific, 2004, pp. 61–141 Search PubMed.
- J. A. Plant, N. Voulvoulis and K. V. Ragnarsdottir, Pollut., Hum. Health Environ., 2012, 181–206 Search PubMed.
- K. Li, X. Zhao, B. K. Hammer, S. Du and Y. Chen, ACS Nano, 2013, 7, 9664–9674 CrossRef CAS PubMed.
- Z. Magdolenova, Y. Lorenzo, A. Collins and M. Dusinska, J. Toxicol. Environ. Health, Part A, 2012, 75, 800–806 CrossRef CAS PubMed.
- L. Lubinski, P. Urbaszek, A. Gajewicz, M. Cronin, S. Enoch, J. Madden, D. Leszczynska, J. Leszczynski and T. Puzyn, SAR QSAR Environ. Res., 2013, 24, 995–1008 CrossRef CAS PubMed.
- Z. Zhao, Y. Huang, B. Zhang, Y. Shyr and H. Xu, BMC Genomics, 2012, 13(8), S1 Search PubMed.
- X.-R. Xia, N. A. Monteiro-Riviere and J. E. Riviere, Nat. Nanotechnol., 2010, 5, 671–675 CrossRef CAS PubMed.
- T. Puzyn, B. Rasulev, A. Gajewicz, X. Hu, T. P. Dasari, A. Michalkova, H.-M. Hwang, A. Toropov, D. Leszczynska and J. Leszczynski, Nat. Nanotechnol., 2011, 6, 175–178 CrossRef CAS PubMed.
- T. Puzyn, D. Leszczynska and J. Leszczynski, Small, 2009, 5, 2494–2509 CrossRef CAS PubMed.
- T. Petrova, B. F. Rasulev, A. A. Toropov, D. Leszczynska and J. Leszczynski, J. Nanopart. Res., 2011, 13, 3235–3247 CrossRef CAS.
- A. Gajewicz, B. Rasulev, T. Dinadayalane, P. Urbaszek, T. Puzyn, D. Leszczynska and J. Leszczynski, Adv. Drug Delivery Rev., 2012, 64, 1663–1693 CrossRef CAS PubMed.
- A. Gajewicz, N. M. Schaeublin, E. I. Maurer, S. M. Hussain, B. Rasulev, D. Leszczynska, J. Leszczynski and T. Puzyn, Nanotoxicology, 2015, 9(3), 313–325 CrossRef CAS PubMed.
- D. Fourches, D. Pu, C. Tassa, R. Weissleder, S. Y. Shaw, R. J. Mumper and A. Tropsha, ACS Nano, 2010, 4, 5703–5712 CrossRef CAS PubMed.
- E. Burello and A. Worth, Nanotoxicology, 2011, 5, 228–235 CrossRef CAS PubMed.
- N. Sizochenko, B. Rasulev, A. Gajewicz, V. Kuz'min, T. Puzyn and J. Leszczynski, Nanoscale, 2014, 6, 13986–13993 RSC.
- M. Turabekova, B. Rasulev, M. Theodore, J. Jackman, D. Leszczynska and J. Leszczynski, Nanoscale, 2014, 6, 3488–3495 RSC.
- L. Ahmed, B. Rasulev, M. Turabekova, D. Leszczynska and J. Leszczynski, Org. Biomol. Chem., 2013, 11, 5798–5808 CAS.
- C.-H. Peng, Y.-Z. Jiang, A.-S. Tai, C.-B. Liu, S.-C. Peng, C.-T. Liao, T.-C. Yen and W.-P. Hsieh, Nucleic Acids Res., 2014, 42, 2803–2819 CrossRef CAS PubMed.
- G. F. Cooper and C. N. Glymour, Computation, causation, and discovery, AAAI Press, MIT Press, Menlo Park, California, 1999 Search PubMed.
- J. Koster, Stat. Med., 2003, 22, 2236–2237 CrossRef PubMed.
- J. Pearl, Causality: models, reasoning, and inference, Cambridge University Press, 2000, pp. 1–384 Search PubMed.
- P. Hoyer, D. Janzing, J. Mooij, J. Peters and B. Schölkopf, presented in part at the Twenty-Second Annual Conference on Neural Information Processing Systems (NIPS 2008), Vancouver, BC, Canada, 2009 Search PubMed.
- P. Spirtes, C. Glymour and R. Scheines, in Causation, Prediction, and Search, Springer, New York, 1993, vol. 81, ch. 5, pp. 103–162 Search PubMed.
- P. Spirtes, C. Glymour and R. Scheines, in Causation, Prediction, and Search, Springer, New York, 1993, vol. 81, ch. 3, pp. 41–86 Search PubMed.
- P. Daniusis, D. Janzing, J. Mooij, J. Zscheischler, B. Steudel, K. Zhang and B. Schölkopf, arXiv preprint arXiv:1203.3475, 2012.
- D. Janzing, J. Mooij, K. Zhang, J. Lemeire, J. Zscheischler, P. Daniu and B. Schölkopf, Artif. Intell., 2012, 182–183, 1–31 CrossRef PubMed.
- H. Zhang, Z. Ji, T. Xia, H. Meng, C. Low-Kam, R. Liu, S. Pokhrel, S. Lin, X. Wang, Y.-P. Liao, M. Wang, L. Li, R. Rallo, R. Damoiseaux, D. Telesca, L. Mädler, Y. Cohen, J. I. Zink and A. E. Nel, ACS Nano, 2012, 6, 4349–4368 CrossRef CAS PubMed.
- A. G. Artemenko, E. N. Muratov, V. E. Kuz'min, N. N. Muratov, E. V. Varlamova, A. V. Kuz'mina, L. G. Gorb, A. Golius, F. C. Hill and J. Leszczynski, SAR QSAR Environ. Res., 2011, 22, 575–601 CrossRef CAS PubMed.
- A. P. Mathews, Am. J. Physiol., 1904, 10, 290–323 CAS.
- C. P. Tatara, M. C. Newman, J. T. McCloskey and W. P. Li, Aquat. Toxicol., 1998, 42, 255–269 CrossRef CAS.
- B. M. Smirnov, Phys.-Usp., 2011, 54, 691–721 CrossRef CAS.
- R. Liu, H. Y. Zhang, Z. X. Ji, R. Rallo, T. Xia, C. H. Chang, A. Nel and Y. Cohen, Nanoscale, 2013, 5, 5644–5653 RSC.
- W. H. Qi and M. P. Wang, Mater. Chem. Phys., 2004, 88, 280–284 CrossRef CAS PubMed.
- V. K. Sharma, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2009, 14, 1485–1495 CrossRef PubMed.
- L. Breiman, Mach. Learn., 2001, 45, 5–32 CrossRef.
- P. G. Polishchuk, E. N. Muratov, A. G. Artemenko, O. G. Kolumbin, N. N. Muratov and V. E. Kuz'min, J. Chem. Inf. Model., 2009, 49, 2481–2488 CrossRef CAS PubMed.
- M. Hohn, Math. Geol., 1976, 8, 137–150 CrossRef.
- Y.-N. Chang, M. Zhang, L. Xia, J. Zhang and G. Xing, Materials, 2012, 5, 2850–2871 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra11399g |
| This journal is © The Royal Society of Chemistry 2015 |





