Porcine circovirus type 2 affects the serum profile of amino acids and intestinal expression of amino acid transporters in mice
Abstract
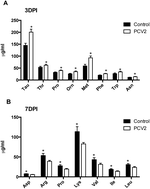
PCV2 is highly pathogenic, however, its effect on the serum amino acids profile is unknown. This study was conducted to explore the profile of amino acids in the serum in porcine circovirus type 2 (PCV2) infected mice. The serum levels of amino acids were detected with isotope dilution liquid chromatography-mass spectrometry methods at 3, 7, 10 and 14 days post infection (DPI). Meanwhile, the expression of seven amino acids transporters (SLC6A14, SLC6A20, SLC7A5, SLC7A6, SLC7A7, SLC7A8, SLC7A9) in the jejunum was analyzed by reverse transcription polymerase chain reaction (RT-PCR) at 3 and 7 DPI. Serum PCV2 load was also analyzed by quantitative PCR at 3, 7, 10 and 14 DPI. Serum levels of most amino acids, such as Pro, Orn, and Met, significantly (P < 0.05) increased at 3 DPI. However, most amino acids, including Asp, Sar, Arg, Hyl, Pro, Lys, Val, Ile and Leu, significantly (P < 0.05) decreased at 7 DPI. There was no significant difference for most amino acids at 10 and 14 DPI. PCV2 infection significantly (P < 0.05) decreased expression of SLC7A5 and SLC7A6 at 7 DPI. In conclusion, PCV2 infection affects the profile of amino acids in the serum and the expression of amino acids transporters in the intestine.

 Please wait while we load your content...
Please wait while we load your content...