A near-infrared chemodosimeter with Pi-selective colorimetric and fluorescent sensing and its application in vivo imaging†
Tie Nan Zanga,
Rui Rui Zhaoa,
Xing Zhu Yanga,
Yan Gaoa,
Guang Ke Wanga,
Ying Zhou*a,
Bo Liub and
Jun Feng Zhang*b
aKey Laboratory of Medicinal Chemistry for Natural Resource, School of Chemical Science and Technology, Yunnan University, Kunming 650091, China. E-mail: yingzhou@ynu.edu.cn
bCollege of Chemistry and Chemical Engineering, Yunnan Normal University, Kunming 650500, China. E-mail: junfengzhang78@aliyun.com
First published on 10th August 2015
Abstract
A near-infrared colorimetric and fluorescent chemosensor for detecting phosphate ion (Pi) has been developed. The chemosensor's sensing mechanism is based on the Pi-driven cleavage of an amide bond which releases a fluorophore. The chemosensor exhibits a rapid response and high sensitivity towards Pi in DMSO–HEPES buffer (0.02 M, pH = 7.0) (v/v = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). A colorimetric change with a 70 nm red-shift in the UV-vis absorbance spectrum and a 72-fold enhancement in the ratio of fluorescence intensities at 656 nm and 552 nm were observed. The practical utility of this chemosensor was demonstrated by employing it to detect Pi in Paramecium and C. elegans.
1). A colorimetric change with a 70 nm red-shift in the UV-vis absorbance spectrum and a 72-fold enhancement in the ratio of fluorescence intensities at 656 nm and 552 nm were observed. The practical utility of this chemosensor was demonstrated by employing it to detect Pi in Paramecium and C. elegans.
In recent years, fluorescence imaging has become one of the most powerful techniques for in vivo monitoring.1–3 So far, a large number of biomolecule-sensing fluorescent probes have been constructed.4,5 ATP (adenosine triphosphate), a star molecule, has been a target of these efforts, owing to its fundamental contributions to energy transduction and involvement in multiple metabolic pathways.6–8 In contrast, the phosphate ion (Pi), an important downstream metabolic product of ATP, has not attract much attention in last decade even though it has also been reported to widely participate in signaling and regulating biological processes.8–10 To the best of our knowledge, very few Pi sensors have been reported with the exception of the series of Pi-sensing chemodosimeters we published very recently.11–14
Most of the fluorescence sensors have absorption and emission peaks in the visible range (400–650 nm). By contrast, relatively few near-infrared (NIR) fluorescent sensors (absorption and emission in the 650–900 nm range) have been reported.15,16 NIR sensors have many advantages over visible-range sensor, such as reduced photodamage to biological samples, deep tissue penetration, and minimal interference from background autofluorescence by biomolecules. Therefore, we set out to develop a highly selective NIR Pi sensor and apply it successfully to in vivo imaging. In signal mode, a ratiometric response was considered as a better choice in our design because they employ the ratio of the emission intensities at two different wavelengths, which could avoid interference from the inhomogeneous distribution of fluorescent indicators in cells.17,18
We successfully designed a reaction-based NIR sensor, 1, which contains an oxalate moiety as a Pi-responsive trigger. In sensor 1, the reaction site, oxalate group, is linked via an amide bond to a derivative of dicyanomethylene-4H-chromene, well-known red emission dye with an ICT characteristic. According to our experience, we anticipated that Pi would lead to the cleavage of amide bond, bringing about marked ratiometric colorful and fluorescence changes.
This work demonstrated that 1 is a selective NIR Pi sensor which does not appreciably react to other phosphate-containing species including PPi, ATP, TTP, AMP, UTP, and GMP, and which is suitable for in vivo imaging. As expected, only the addition of Pi brought about marked color and fluorescence changes in solutions of 1 (DMSO–HEPES buffer, 0.02 M, pH = 7.0, v/v = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). A dramatic solution change from yellow to orange, with a 70 nm red-shift in the UV-vis spectrum and an appearance of a new NIR peak at 664 nm were observed. An 72-fold enhancement in the ratio of fluorescence intensities at 656 nm and 552 nm (F656/F552) ensure 1 as a ratiometric NIR fluorescence sensor. The probe is also successfully applied to visualize exogenous and endogenous Pi in paramecium and C. elegans.
1). A dramatic solution change from yellow to orange, with a 70 nm red-shift in the UV-vis spectrum and an appearance of a new NIR peak at 664 nm were observed. An 72-fold enhancement in the ratio of fluorescence intensities at 656 nm and 552 nm (F656/F552) ensure 1 as a ratiometric NIR fluorescence sensor. The probe is also successfully applied to visualize exogenous and endogenous Pi in paramecium and C. elegans.
Probe 1 was successfully synthesized in one step starting from compound 2, which was synthesized as described previously.19–21 As shown in Scheme 1, compound 2 reacted with ethyl oxalyl chloride in the presence of triethylamine (TEA) in dry dichloromethane at room temperature, giving the target compound 1 with a yield of 35%. Experimental details and corresponding NMR spectra and mass spectra are summarized in the ESI.†
In order to study the selectivity of probe 1 for Pi ions, spectral changes (UV/vis and fluorescence) of 1 were monitored in the presence of various anions (as sodium salts) in DMSO–HEPES buffer. Absorption and emission spectra were acquired 3 min after addition of anions into the probe solutions. The UV-vis absorbance spectrum of 1 in DMSO–HEPES buffer (pH = 7.0) contains a peak located at 448 nm. As expected, the absorption spectrum of 1 undergoes a dramatic change only when Pi is added. The addition of Pi led to the reduction of the major absorbance peak at 448 nm with one new absorption peak appearing at 518 nm. When other anions and molecules (P2O74−, GSH, ATP, GMP, TTP, UDP, Pi, TDP, L-Glu, NO2−, S2O32−, S2O52−, SO42−, NO3−, Cys) were added at the same levels as Pi (120 equiv.), negligible spectral changes were observed (Fig. 1).
As demonstrated in Fig. 2, successive addition of Pi into the probe solution of 1 in DMSO–HEPES (0–120 equiv.), led to a decrease in the absorption at 448 and a simultaneous increase in a new peak at 518 nm, with an isoabsorptive point at 464 nm. As predicted, the solution color undergoes a dramatic change when 1 is incubated with 120 equiv. of Pi. The solution changes from yellow to orange, with a 70 nm red-shift in the spectrum. These results indicate that 1 could serve as a “nake-eye” probe for Pi with high selectivity.
 | ||
Fig. 2 Absorbance emission titration spectra of 1 (3.0 × 10−5 M) in the presence of varying concentrations of Pi in DMSO–HEPES buffer (0.02 M, pH = 7.0) (v/v = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1). | ||
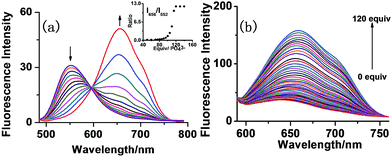
The results of fluorescence spectroscopy show that 1 in DMSO–HEPES buffer (0.02 M, pH = 7.0) (v/v = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) emits with a maximum at 552 nm upon excitation at 450 nm. (Fig. 3a) Only the addition of 120 equiv. Pi to the solution of 1 caused a simultaneous decrease in the intensity of band at 552 nm and increase in a new band at 656 nm. All the other tested anions could not lead to the increase of the band at 656 nm, as shown in Fig. 4. Concentration dependent titration study showed that in the presence of Pi, a decrease at 552 nm and increase at 656 nm take place simultaneously and reach respective minimum and maximum values at a 120 equiv. of Pi, with a desirable ratiometric type probe feature. It showed that the ratio of fluorescence intensities at 656 nm and 552 nm (F656/F552) increased linearly with increasing Pi concentration, resulting in a 71-fold enhancement. A detection limit as low as 1.36 × 10−6 M was established by using 1 with a signal-to-noise ratio of 3 (Fig. S3†).
1) emits with a maximum at 552 nm upon excitation at 450 nm. (Fig. 3a) Only the addition of 120 equiv. Pi to the solution of 1 caused a simultaneous decrease in the intensity of band at 552 nm and increase in a new band at 656 nm. All the other tested anions could not lead to the increase of the band at 656 nm, as shown in Fig. 4. Concentration dependent titration study showed that in the presence of Pi, a decrease at 552 nm and increase at 656 nm take place simultaneously and reach respective minimum and maximum values at a 120 equiv. of Pi, with a desirable ratiometric type probe feature. It showed that the ratio of fluorescence intensities at 656 nm and 552 nm (F656/F552) increased linearly with increasing Pi concentration, resulting in a 71-fold enhancement. A detection limit as low as 1.36 × 10−6 M was established by using 1 with a signal-to-noise ratio of 3 (Fig. S3†).
The sensing mechanism of 1 was proven by the tests of 1H NMR titration (Fig. S5†) and HRMS (Fig. S6†), and the proposed sensing mechanism was shown in Scheme 2. According to our prior work,11–14 Pi initially attacks the carbon atom of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and subsequently reacts with the carbon atom of C–N,22–25 resulting in the decomposition of 1 into 2 and 6. Therefore, the NIR optical properties of 2 was final released and shown. The 1H NMR titration spectra of compound 1 (0.5 mM) with Pi in DMSO-d6 was measured. It showed that final product of the sensing process of Pi by 1 was 2 (Fig. S5†). Meanwhile, the HRMS of the mixture of 1 and Pi was tested after the two compounds were mixed over night. According to the data, the peak of 2 became the main peak while that of 1 disappeared. All the data gave the strong supports for the sensing mechanism as we expected.
O, and subsequently reacts with the carbon atom of C–N,22–25 resulting in the decomposition of 1 into 2 and 6. Therefore, the NIR optical properties of 2 was final released and shown. The 1H NMR titration spectra of compound 1 (0.5 mM) with Pi in DMSO-d6 was measured. It showed that final product of the sensing process of Pi by 1 was 2 (Fig. S5†). Meanwhile, the HRMS of the mixture of 1 and Pi was tested after the two compounds were mixed over night. According to the data, the peak of 2 became the main peak while that of 1 disappeared. All the data gave the strong supports for the sensing mechanism as we expected.
Paramecium is a slipper shaped ciliate which is found in oxygenated aquatic environments feeding near vegetative matter.26 This organism is quite common and may easily be obtained from the shallow waters. Paramecia are a key link in detritus-based food webs in aquatic ecosystems. Most paramecia are feed voraciously on bacteria that accompany decaying organic matter. These bacteria-gorged cells are then consumed by other protists and small animals, which are in turn preyed upon by larger organisms.27 Despite its small size, a paramecium has a relative structure compared with single cells. To follow the aggregation of the endogenous and the exogenous Pi in paramecium attracted our attention. For this purpose, paramecia were incubated with 5 mM Pi for 12 h first. The paramecia were observed to display red fluorescence after the incubation of Pi.
Apyrase, a hydrolytic enzyme that converts both ATP and ADP into AMP and Pi, was utilized to evaluate the ability of 1 to measure in vivo endogenous Pi production. In a second group, paramecia were treated with ATP, and then various amounts of apyrase were added to the test solutions. To paramecia preincubated with ATP (1 mM) for 2 h at 37 °C, apyrase was added (0, 1, 5, and 10 U), followed by staining with probe 1 for 2 h. As shown in Fig. 5, no fluorescence could be observed when paramecia were treated only in the presence of ATP. With the addition of apyrase, the fluorescence intensity increased as the concentrations of apyrase increased. The brightest red fluorescence was found with 10 U apyrase. Furthermore, bright fluorescence was observed in both the micronucleus and macronucleus.
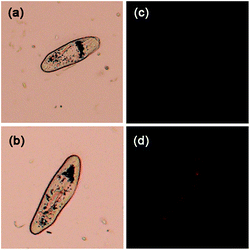 | ||
| Fig. 5 Fluorescent imaging (right) and phase contrast (left) for Pi in paramecium (a) and (b): bright field. (c) Probe 1 (20 μM). (d) Probe 1 (20 μM) and Pi (1 mM). | ||
Finally, the use of 1 with fluorescence microscopy to track Pi inside C. elegan larvae at developmental stage 4 (L4) was explored. (Fig. 7) Yellow fluorescence was observed in the nematodes, pretreated with 1 (10 μM) for 2 h. When the Pi was added, a bright red fluorescence is emitted from the whole body of nematodes. These combined results indicated that 1 is cell membrane permeable and that it may be employed as an NIR fluorescence imaging agent for the detection of Pi in both paramecia and C. elegans.28,29
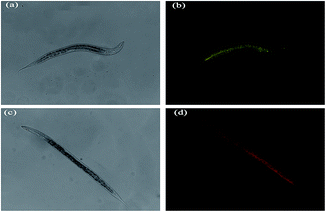 | ||
| Fig. 7 Fluorescent imaging (top) and phase contrast (bottom) for Pi in C. elegans. (a) and (c): bright field. (b) Probe 1 (20 μM) only. (d) Probe 1 (20 μM), Pi (1 mM). | ||
Conclusions
In conclusion, a functional NIR fluorescent chemodosimeter 1, bearing an oxalate moiety as a Pi responsive trigger, was developed. The optical properties of 1 were tested, and demonstrated that this sensor has high selectivity and excellent sensitivity towards Pi. The chemosensor exhibited a remarkable colorimetric change and ratiometric fluorescence enhancement in response to Pi. Furthermore, 1 was proven to be capable of imaging endogenously produced Pi as well as exogenous Pi in living Paramecium and C. elegans, demonstrating the value of our compound as an in vivo NIR fluorescent probe.Acknowledgements
This work was supported by the Natural Science Foundation of China (21262045, 21262050, 21302165, and 21462050), the Foundation of the Department of Science and Technology of Yunnan Province of China (2013HB062, 2014HB008), and training project (XT 412004) of Yunnan University.Notes and references
- X. Wang, L. Cui, N. Zhou, W. Zhu, R. Wang, X. Qian and Y. Xu, Chem. Sci., 2013, 4, 2936 RSC.
- Y. Zhou, J. F. Zhang and J. Yoon, Chem. Rev., 2014, 114, 5511–5571 CrossRef CAS PubMed.
- K. P. Divya, S. Sreejith, P. Ashokkumar, K. Yuzhan, Q. Peng, S. K. Maji, Y. Tong, H. Yu, Y. Zhao, P. Ramamurthy and A. Ajayaghosh, Chem. Sci., 2014, 5, 3469–3474 RSC.
- X. J. Zou, Y. C. Ma, L. E. Guo, W. X. Liu, M. J. Liu, C. G. Zou, Y. Zhou and J. F. Zhang, Chem. Commun., 2014, 50, 13833–13836 RSC.
- Y. Zhou and J. Yoon, Chem. Soc. Rev., 2012, 41, 52–67 RSC.
- P. Mahato, A. Ghosh, S. K. Mishra, A. Shrivastav, S. Mishra and A. Das, Inorg. Chem., 2011, 50, 4162–4170 CrossRef CAS PubMed.
- Y. Zhou, Z. Xu and J. Yoon, Chem. Soc. Rev., 2011, 40, 2222–2235 RSC.
- R. K. Pathak, V. K. Hinge, A. Rai, D. Panda and C. P. Rao, Inorg. Chem., 2012, 51, 4994–5005 CrossRef CAS PubMed.
- S. Khoshniat, A. Bourgine, M. Julien, P. Weiss, J. Guicheux and L. Beck, Cell. Mol. Life Sci., 2011, 68, 205–218 CrossRef CAS PubMed.
- A. S. Rao, D. Kim, H. Nam, H. Jo, K. H. Kim, C. Ban and K. H. Ahn, Chem. Commun., 2012, 48, 3206–3208 RSC.
- L. E. Guo, J. F. Zhang, X. Y. Liu, L. M. Zhang, H. L. Zhang, J. H. Chen, X. G. Xie, Y. Zhou, K. Luo and J. Yoon, Anal. Chem., 2015, 87, 1196–1201 CrossRef CAS PubMed.
- H. Wang, L. E. Guo, X. M. Li, L. M. Zhang, Q. L. Xu, G. F. Wu, Y. Zhou and J. F. Zhang, Dyes Pigments, 2015, 120, 293–298 CrossRef CAS PubMed.
- J. F. Zhang, L. E. Guo, T. N. Zang, Y. L. Duan, X. Y. Liu, Z. G. Yang, P. Verwilst, K. J. Luo, G. K. Wang, J. F. Kou, Y. Zhou and J. S. Kim, Chem.–Asian J., 2015, 10, 1165–1169 CrossRef CAS PubMed.
- G. F. Wu, Q. L. Xu, L. E. Guo, T. N. Zang, R. Tan, S. T. Tao, J. F. Ji, R. T. Hao, J. F. Zhang and Y. Zhou, Tetrahedron Lett., 2015, 56, 5034–5038 CrossRef CAS PubMed.
- L. Yuan, W. Lin, Y. Yang and H. Chen, J. Am. Chem. Soc., 2012, 134, 1200–1211 CrossRef CAS PubMed.
- A. Matsui, K. Umezawa, Y. Shindo, T. Fujii, D. Citterio, K. Oka and K. Suzuki, Chem. Commun., 2011, 47, 10407–10409 RSC.
- J. Wang, F. Song, J. Wang and X. Peng, Analyst, 2013, 138, 3667–3672 RSC.
- A. Ajayaghosh, P. Carol and S. Sreejith, J. Am. Chem. Soc., 2005, 127, 14962–14963 CrossRef CAS PubMed.
- W. Sun, J. Fan, C. Hu, J. Cao, H. Zhang, X. Xiong, J. Wang, S. Cui, S. Sun and X. Peng, Chem. Commun., 2013, 49, 3890–3892 RSC.
- W. Zhu, X. Huang, Z. Guo, X. Wu, H. Yu and H. Tian, Chem. Commun., 2012, 48, 1784–1786 RSC.
- Z. Guo, W. Zhu and H. Tian, Chem. Commun., 2012, 48, 6073–6084 RSC.
- X. Wu, X. Sun, Z. Guo, J. Tang, Y. Shen, T. D. James, H. Tian and W. Zhu, J. Am. Chem. Soc., 2014, 136, 3579–3588 CrossRef CAS PubMed.
- B. Tang, Y. Xing, P. Li, N. Zhang, F. Yu and G. Yang, J. Am. Chem. Soc., 2007, 129, 11666–11667 CrossRef CAS PubMed.
- J. Bouffard, Y. Kim, Y. M. Swager, R. Weissleder and S. A. Hilderbrand, Org. Lett., 2007, 10, 37–40 CrossRef PubMed.
- M. M. Pires and J. Chmielewski, Org. Lett., 2007, 10, 837–840 CrossRef PubMed.
- R. Wichterman, The Biology of Paramecium, Plenum Press, New York, 1986 Search PubMed.
- L. Margulis, and K. V. Schwartz, Five Kingdoms-An Illustrated Guide to the Phyla of Life on Earth, W.H. Freeman, New York, 3rd edn, 1998 Search PubMed.
- L. M. Zhang, L. E. Guo, X. M. Li, Y. G. Shi, G. F. Wu, X. G. Xie, Y. Zhou, Q. H. Zhao and J. F. Zhang, Tetrahedron Lett., 2014, 55, 6131–6136 CrossRef CAS PubMed.
- G. K. Wang, Q. L. Mi, L. Y. Zhao, J. J. Hu, L. E. Guo, X. J. Zou, B. Liu, X. G. Xie, J. F. Zhang, Q. H. Zhao and Y. Zhou, Chem.–Asian J., 2013, 9, 744–748 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic detail, crystal data. CCDC 1032275, job plot, detecting limiting and spectroscopic data. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c5ra12835h |
| This journal is © The Royal Society of Chemistry 2015 |