Pd nanoparticles supported on reduced graphene–E. coli hybrid with enhanced crystallinity in bacterial biomass†
Abstract
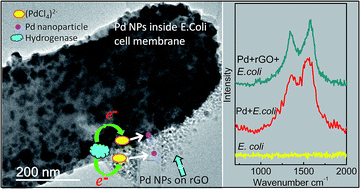
A novel method for simultaneous reduction of graphene oxide (GO) and palladium salt, Pd(II), using Escherichia coli (E. coli) in the separate presence of two different mild reducing agents (hydrogen and formate) is investigated to successfully produce reduced GO (rGO)-biomass/Pd hybrid material for potential use as an electrocatalyst. Transmission electron microscopy, X-ray diffraction, thermo-gravimetric analysis, X-ray photoelectron spectroscopy and Raman microscopy demonstrate the successful reduction of Pd(II), GO and the biomass, resulting in the formation of Pd nanoparticles (PdNPs) on an rGO–biomass hybrid. The distribution of the NPs was found to be dependent on the type of reducing agent. PdNPs formed on rGO sheets showed relatively uniform distribution and size control (2–5 nm), whereas PdNPs on the bacterial scaffold were larger (up to 10 nm in size). Raman spectroscopy studies suggest that the presence of Pd leads to oxygen reduction and increased crystallinity in the bacterial biomass. Previous studies have suggested the potential for a bacterially-supported Pd electrocatalyst in fuel cells and, independently, the suitability of rGO as a support for PdNPs. This study confirms the simultaneous bacterial reduction of Pd(II) and GO and the association between the bacterial cells and rGO. We suggest that the simultaneous presence of E. coli and mild reducing agent together with GO and Pd(II) creates an interactive and synergistic environment in a hybrid material to allow (a) better control of PdNP size and distribution both on the inside of the bacterial membrane and on the rGO sheets and (b) increased crystallinity of the bacterial biomass compared to systems with bacteria alone.

 Please wait while we load your content...
Please wait while we load your content...