Distinguishing the serum metabolite profiles differences in breast cancer by gas chromatography mass spectrometry and random forest method
Abstract
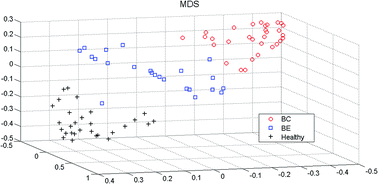
In this study, we proposed a metabolomics strategy to distinguish different metabolic characters of healthy controls, breast benign (BE) patients, and breast malignant (BC) patients by using the GC-MS and random forest method (RF). In the current study, the serum samples from healthy controls, BE patients, and BC patients were characterized by using GC-MS. Then, random forest (RF) models were established to visually discriminate the differences among three groups' metabolites profiles, and further investigate the progress of breast cancer from benign to malignant in patients based on these GC-MS profiles. We successfully discovered the differences between the healthy and breast cancer patients. And the metabolic changes from benign to malignant cancer were obviously visualized. The results suggested that combining GC-MS profiling with random forest method is a useful approach to analyze metabolites and to screen the potential biomarkers for exploring the serum metabolic profiles of breast cancer.

 Please wait while we load your content...
Please wait while we load your content...