Open Access Article
Open Access ArticleConditioned bio-interfaces of silicon/porous silicon micro-patterns lead to the chondrogenesis of hMSCs
H.
Alarcón
a,
M. D.
Ynsa
*bc,
Z. Y.
Dang
d,
V.
Torres-Costa
bc,
M.
Manso-Silván
b,
J. F.
Wu
d,
M. B. H.
Breese
d and
J. P.
García-Ruiz
a
aMolecular Biology Department, Universidad Autónoma de Madrid, Campus de Cantoblanco, 28049 Madrid, Spain
bDepartment of Applied Physics and Instituto Nicolás Cabrera, Universidad Autónoma de Madrid, Campus de Cantoblanco, 28049 Madrid, Spain. E-mail: m.ynsa@uam.es
cCentro de Micro-Análisis de Materiales (CMAM), Universidad Autónoma de Madrid, Campus de Cantoblanco Edif, 22, Faraday 3, E-28049 Madrid, Spain
dCentre for Ion Beam Applications (CIBA), Department of Physics, National University of Singapore, Singapore 117542
First published on 6th October 2015
Abstract
The interaction between cells and materials is of scientific and technological interest for the development of new biomaterials with improved functional properties. In this work the chondrogenesis (CG) of human mesenchymal stem cells (hMSCs) derived from the bone marrow of healthy donors has been achieved on Si/PSi surfaces after pre-treatment with a conditioned medium derived from hMSCs (CM). The chondrogenic process was analyzed in fixed-cell preparations by immune fluorescence, determining the cellular localization of β-catenin and the transcription factors STAT-5, Runx2 and vitamin D receptor (VDR). In a week of CG on Si/PSi or CM-Si/PSi, β-catenin showed cell–cell contacts priming a preferential chondrogenic process on a PSi surface, in contrast with the preferential proliferation and migration processes observed on hexagonal/triangular micro-designed Si areas. Interestingly, the presence of a CM bio-interface improves this differentiation process with respect to a control PSi surface. The chondroinductive effect was also observed in STAT-5, which was highly expressed and is involved in intracellular signaling of many differentiation receptors related to development and tissue repair. In good agreement with the above results, the co-localization of transcription factors Runx2/VDR at nuclei supports active transcription of chondrogenic genes. Considering these observations, we propose that the combination of a rough PSi surface and the components of the CM-hMSCs bio-interface boosts the chondrogenesis of hMSCs. These findings suggest that the tailoring of Si/PSi scaffolds with appropriate bio-interface combinations improves their therapeutic potential.
1. Introduction
One of the main goals of tissue engineering is the interaction of cells and tissues with biomaterial surfaces to preserve and control specific biological processes involved in tissue repair and disease reversal. Tissues have a variable capacity to repair/regenerate, such as in the liver, blood and to a limited extent in bone or cartilage. Although the chondrogenesis process for repairing hyaline cartilage during adulthood is very poor, skeletal tissues derived from endochondral ossification of mesenchymal stem cell (MSC) aggregates maintain this process. Such events can be observed from skeletal development to the remodeling/repair of the pelvis, in bone extremities and in the vertebral column throughout life.1 This complex process requires a different cellular mechanism, which is mediated by cell-condensation and differentiation, carried out in a bone-like niche composed of adequate factors, chemical and physical conditions. However, the cellular stages at which MSCs are supposed to transform from progenitors to differentiated skeletal tissues and the molecular mechanisms involved are not yet well understood.Chondrogenesis (CG) is initiated when MSCs stop dividing and begin to express extracellular matrix components (ECM), such as protein factors, proteoglycans and collagens, forming the niche for MSC-derived cells. The bone marrow niche is indispensable also for hematopoietic stem cells that differentiate in progenitor cells of the immune/hematopoietic system, as well as for endothelial cells to form and maintain vascular health. Cells recognize the ECM geometry in order to regulate intracellular architecture and to provide spatial information on cell polarization, the stability of intercellular junction locations and the magnitude of intra and intercellular forces.2,3 The heterogeneity of the spatial organization of ECM induces an anisotropic distribution of mechanical constraints in cells at the time they adapt to minimize both intra- and intercellular forces. The exact contribution of cell–cell and cell–ECM interactions to the specific homeostasis of a niche area is starting to be studied. Increasing amounts of data suggest that MSCs reside in a perivascular niche where they communicate, producing soluble factors and components of ECM.4 The use of these mixtures of soluble factors, extracellular proteoglycans and collagen produced by hMSCs, known as conditioned medium (CM-hMSCs), has been shown to be effective in forming a bio-interface that increases the adhesion of hMSCs,5–8 including sponge structures of PDMS.9
The concept of a scaffold has changed from being considered an inert material to being a proactive cell-instructive material able to control and guide morphogenesis and tissue repair, by displaying the biochemical and biophysical processes needed for tissue formation, maintenance and function. It is known that changes in material properties such as hydrophobicity, surface charge, roughness and stiffness are important for cell-response.10 Si in the form of porous silicon (PSi) is an extremely versatile material and displays a wide range of surface properties depending on processing, aging and functionalization routes.11,12 A way to visualize the contrasting attraction of PSi towards cells consists in the integration of its different forms into micro-patterned surfaces.13,14 In this study, we assess whether a micro-patterned Si/PSi surface can induce chondrogenesis and whether pretreatment with CM-hMSCs could generate a bio-interface that improves this process.
2. Materials and methods
2.1 Description and preparation of Si/PSi micro-patterns
The micro-patterned Si/PSi surfaces employed in this work are similar to those used in previous studies on hMSC growth and migration.14 As previously described, these surfaces consist of a central Si regular hexagon (side length 5 mm) surrounded by tracks (2 mm length) of Si triangles of different sizes (triangle size 10 μm, 25 μm or 50 μm), and all Si regions are surrounded by PSi areas with pore diameters of a few tens of nanometers. These micro-patterns were precisely fabricated by a non-conventional lithography method for biological purposes. Bulk p-type Si wafers were selectively irradiated with a high energy ion beam (500 keV H2+). The ion-induced defects locally reduce the free carrier density, hence increasing the resistivity of the irradiated silicon regions. Consequently, during the subsequent electrochemical anodisation, the PSi formation rate is reduced in the irradiated areas compared to that in the non-irradiated regions, and the surface roughness is also different.2.2 Cell culture and chondrogenesis
Two to four milliliters of human bone marrow samples from healthy donors were used to isolate hMSC as described.15 hMSCs were provided by Dr Benjamin Fernández from the Foundation Jiménez-Diaz (Madrid, Spain) and the cell-bank of Málaga University (Málaga, Spain). The culture expansion of hMSCs was carried out as previously described.15,16 Culture media were prepared by the Research Service of Molecular Biology Centre Severo Ochoa (CBM, CSIC-UAM). Cells were plated (Falcon) and incubated with DMEM-LG plus 10% FBS of selected batches to 80% confluence. Cells were collected by treatment with 0.25% trypsin–EDTA and seeded at 1.5 × 104 on the central Si hexagon of the Si/PSi micro-patterns. Material surfaces were first washed with Phosphate Buffered Saline (PBS) and sterilized by UV irradiation, then washed and equilibrated using 2 mL of PBS four to five times. As a control, cells were seeded on cover slips coated with 0.5% gelatin (bovine skin, Sigma). Each experiment was realized in duplicate samples and the experiments were repeated 2–3 times. Osteochondral differentiation of hMSC was undertaken following the methodology used in previous works.15–19 Cells were incubated with 2 mL of DMEM-LG with ITS (6.25 μg mL−1 insulin; 6.25 μg mL−1 transferrin; 6.25 μg mL−1 selenous acid) (Collaborative Research), 400 μM AANE (non-essential amino acids, CBM), 2 mM pyruvate (Gibco), 37.5 μg mL−1 ascorbate (WAKO), 0.1 mM dexamethasone (Decadran, Merck), 1 mM β-glycerol, and 0.06 ng mL−1 TGF-b3 (R&D System) for one week. The medium was replaced twice per week.2.3 hMSCs conditioned medium
For the preparation of each CM-hMSCs batch, 8–10 p100 culture plates of hMSCs grown at 80% confluence in DMEM-LG and 10% FBS were used. Cells were washed thoroughly with PBS and then incubated in DMEM-LG starved of FBS and complemented with 2 mM pyruvate during the following 24 hours. Afterwards the culture medium was collected and cleaned of any floating cells in a bench centrifuge at 1500 rpm for 5 min. The clean supernatant was cooled down in ice for 30 minutes, centrifuged at 9000 rpm in a Sorvall centrifuge for 30 minutes to remove salt precipitations, and the clean supernatant was stored in 2 mL aliquots at −30 °C until use. We avoided repeatedly freeze-thawing the samples. The activity of each batch was measured by wound-healing assay in triplicate using DMEM-LG-Pyr medium and DMEM-LG-10% FBS as controls.2.4 Immunofluorescence
After cell culturing, samples were processed as previously described.20,21 Briefly, culture micro-patterns were rinsed twice with ice-cold PBS and fixed in 3.7% formaldehyde in PBS for 30 min at room temperature (RT), then washed in PBS. Cells were incubated with 0.5% Triton X-100 in CSK (cytoskeleton) buffer containing 10 mM pipes (pH 6.8), 3 mM MgCl2, 100 mM NaCl, 1 mM EGTA, 0.3 M sucrose, and 1 mM PMSF for 30 min on ice to permeate and remove all cell soluble proteins, and fixed with 3.7% formaldehyde as above. For immune detection, cell preparations were blocked with 3% BSA-0.1% Triton X-100 in PBS (PBSA) for 2 h at RT and exposed to different antibodies. We used anti-Runx2 (M-70 and C-19), anti-VDR (D-6 and C-20), anti-STAT-5 (G-2) and anti-β-catenin antibodies (Santa Cruz Biotech Europe), at the concentration indicated by the respective manufacturer. Secondary antibodies were labeled with either Alexa 488 or Alexa 594 (Invitrogen) (Microscopy Service of Molecular Biology Centre, CSIC-UAM, Spain). These antibodies were used at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution. When required, nuclei were stained with DAPI (CALBIOCHEM) for 5 min. Immune staining was observed with an inverted IX81 Olympus associated with a DP72 digital camera and controlled by cellD software.
500 dilution. When required, nuclei were stained with DAPI (CALBIOCHEM) for 5 min. Immune staining was observed with an inverted IX81 Olympus associated with a DP72 digital camera and controlled by cellD software.
2.5 Analysis of data
To determine the cellular expression of β-catenin and STAT-5 in the osteochondral differentiation on native Si/PSi and Si/PSi with CM-hMSCs, we took random fields of the immune staining samples at the same magnification and exposure time using the cellD software. Then, images were processed with ImageJ software to measure the amount of fluorescence corresponding to β-catenin and STAT-5. The obtained values were divided by the number of cells in each field to calculate the amount of β-catenin and STAT-5 per cell (amount of fluorescence per field/no. of cells per field). Due to the limited number of patterned samples, the number of cell lines used in this study was one, which excludes the application of any statistical test. Thus, we decided to show the replicas of the data in boxplots using the statistical computing R program.3. Results and discussion
3.1 Si/PSi and CM-Si/PSi interfaces increase β-catenin in cell–cell adhesion, priming CG of hMSCs
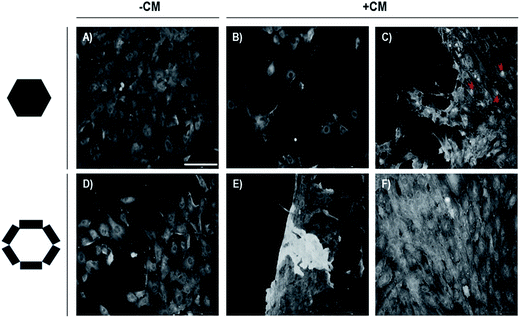
It has been well-demonstrated that CG of hMSCs is induced by incubating micro-masses of cells in a chemically defined medium for three weeks.15,19 Under these conditions, the chondrocytes obtained showed the expression of genes of skeletal pattern, osteochondral differentiation and some protein from ECM.21 The hMSCs plasticity and their communication with ECM have been recognized to be dependent on the patterning of the accompanying tissue. Thus, we assessed whether Si/PSi surfaces could support the CG process of hMSCs, whether there is a preference either for Si or PSi surfaces, and whether there is any alteration of this process by interfacing with CM-Si/PSi. To this end, CG was followed by determining β-catenin complexes as indicative of an integral component of adherent junctions and of the activation of the Wnt pathway. Its absence reflects the inactivation of cadherin cell adhesion and allows cellular proliferation and migration processes, which have clinical relevance associated with cancer development and metastasis.22 The results obtained were gathered both by examining the micrographs in Fig. 1 (A–C for the central hexagonal Si area and D–F for the Si/PSi track areas) and the corresponding replicas of data distribution in the boxplot (Fig. 2). It is observed that β-catenin is highly expressed in hMSCs and localized in the cytoplasm, at cell–cell adhesion conformations on the Si hexagon (A) and cell–PSi adhesions on micro-patterned Si/PSi. Interestingly, when CM-Si/PSi bio-interfaces were obtained, we noted that cell–cell adhesion on Si (B, central and C, border of hexagon) decreased, while it increased on PSi, which showed buried micro-patterns (F) and sporadic detachment at the surface border (E). These results indicate that both PSi surfaces and CM-PSi bio-interfaces are cell-instructive surfaces that can induce the differentiation of hMSCs chondrogenesis. The experimental data have scientific and technological interest for modified scaffolds using a bio-interface that directs specific biological processes.7,8 | ||
| Fig. 2 Boxplots showing data distribution of the β-catenin expression per cell during hMSCs osteochondrogenesis on pristine (−CM) and CM-hMSCs treated (+CM) Si/PSi micropatterns. | ||
3.2 PSi and CM-PSi regulate STAT-5 and VDR
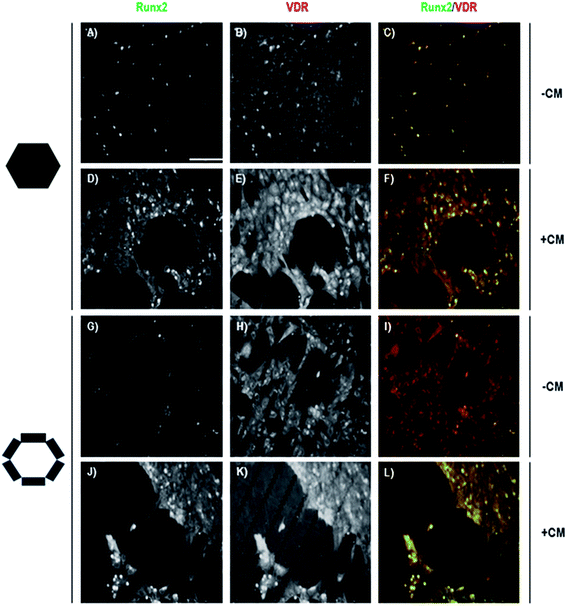
Many factors realize their cellular effect by association to their receptor and activation of the Jak-STATs cellular signaling pathway. Among them are prolactin, growth hormone, erythropoietin, leptin and leukemia inhibitory factor, being essential for energy homeostasis. The STAT-5 member of the STAT family of transcription factors targets various tissues, such as liver, and cell types present in different microenvironments, such as the niches of HSCs (Hematopoietic Stem Cells) and HPCs (Hepatic Progenitor Cells).23,24 We also considered that it would be of interest to analyze the Vitamin D Receptor (VDR) transcription factor member of the nuclear receptor family related with the CG process. As shown in Fig. 3, both STAT-5 and VDR proteins are expressed in cells showing the CG process on a Si/PSi native surface or a CM-Si/PSi bio-interface. In non-pretreated surfaces, cells appeared to be located on Si hexagons with STAT-5 at the cytoplasm (A). As could be expected, VDR was located at the cytoplasm and at nuclei (B) however, no co-localization between both transcription factors could be detected (C). Cells on Si/PSi micro-patterns showed no difference with cells at hexagons (G–I). Interestingly, in a CM-Si/PSi bio-surface an increase in STAT-5 was observed on PSi (J), although, as a result of non-activation by the specific ligand–receptor system, the protein mainly remains at the cytoplasm. VDR, as a member of the nuclear factor family, is also highly expressed on CM-Si/PSi (E), with a preference for PSi (K). Intracellular VDR can be observed in both nuclei (yellow) and cytoplasm. Interestingly, CM treatment did not change any co-localization between both transcription factors (Fig. 3(L) and 4). These results point to CM causing a niche-like inductive CG. Furthermore, it can be suggested that the CM-interfaces retain an adequate proportion of factors that trigger the Jak2-STAT-5 signaling pathway involved in the CG of hMSCs, and direct the entry of cells into G0.25 | ||
| Fig. 4 Boxplots showing data distribution of the STAT-5 expression per cell during hMSCs osteochondrogenesis on pristine (−CM) and CM-hMSCs treated (+CM) Si/PSi micropatterns. | ||
3.3 CM-hMSCs increases nuclear Runx2/VDR co-localization on PSi surface
We also assessed the determination of transcription proteins implicated in bone cell specification. Runx2 and its co-localization with VDR indicate differentiation of hMSCs, since both transcription factors are localized at the gene promoters of cartilage and bone proteins, including the osteocalcin gene.26–31 The results are shown in Fig. 5 and in the boxplot of Fig. 6. As expected from Runx2 and VDR determination, the CM bio-interface increases cell–cell adhesion and the preference for PSi (D, E and J, K) with respect to −CM (A, B, and G, H). Interestingly, this is in agreement with the above results predicted from the catenin and STAT-5 and VDR data. The CM-Si/PSi bio-interface caused a significant increase in the number of cells with nuclear Runx2/VDR co-localization and an improvement on PSi containing track areas. These results fit well with previous works4,32,33 and add a new example to support the important role of physical properties, such as roughness and stiffness of a biomaterial, in allowing progenitor cells to realize two different processes: proliferation/migration and differentiation. Thus, much work needs to be done to improve scaffolds supporting specific fluid dynamics, adequate stiffness and roughness to mimic the desired tissue by directing the scaffold function with bio-interfaces, such as those exemplified herein. | ||
| Fig. 6 Boxplots showing data distribution of the Runx2 expression per cell during hMSCs osteochondrogenesis on pristine (−CM) and CM-hMSCs treated (+CM) Si/PSi micropatterns. | ||
4. Discussion
The results of this work provide data showing that micro-patterned Si/PSi and treatment of these surfaces with CM-hMSCs give rise to bio-interfaces that are inductive scaffolds for hMSCs chondral differentiation. Although the exact composition of this interface is unknown, the results can be explained by the generation of a niche-like surface for bone marrow-derived hMSC adhesion that accelerates the chondral-derived differentiation process. hMSCs seeded on Si/PSi present activation by pretreatment with CM-hMSCs. PSi leads to the expression of the main transcriptional factors for endochondral bone formation. Increased chondrogenesis is also supported by the expanded round morphology of cells, and cell–cell adherence junctions. Furthermore, Runx2/VDR supports chondral differentiation that correlates well with osteocalcin, through which the skeleton exerts the regulation of the whole energetic metabolism of individuals.29,31 Furthermore, increased expression of STAT-5 and its localization at the cytoplasm suggests a traffic of signals between nuclei and cytoplasm involved in the transcriptional networks controlling differentiation in HSCs and progenitor cells.34–36 Interestingly, it has been shown that the STAT-5 transcription factor has a metastasis-suppressive role in breast cancer by inhibiting cancer cell migration, inducing entry into G0, invasion, matrix metalloproteinase secretion and up-regulation of surface E-cadherin and β-catenin.25The data indicate that hMSCs conserve their cellular plasticity on Si, Si/PSi and Si/PSi treated with CM-hMSCs. More precisely, hMSCs grow/migrate on Si and support endochondral bone differentiation on PSi, with both being accelerated on Si/PSi treated with CM-hMSCs. Thus, scaffolds for skeletal-derived tissue could be improved by pretreatment with an adequately conditioned medium. Such a medium is a safe approach to tissue repair, although the precise composition required to induce an extracellular matrix adapted to a particular injured location is not yet known. Many attempts have been made to ascertain the composition of CM-hMSCs for bone regeneration by mass spectrometry and cell expression determination. However, it is a very complex process, starting with the nature of progenitor cells and the jungle of factors that they produce and their relative proportions. No results exist about the specific mixture of ECM components present in CM-hMSCs, neither is there any knowledge of what components are retained directly or indirectly on a particular biomaterial surface.
Being conscious of the complexity of the bone progenitor cell decision, our results support the hypothesis that PSi treated with CM-hMSCs constitutes a bio-active interface directing the cartilage and endochondral bone formation of hMSCs. These results may be taken into account when considering new possible therapies for osteoporosis, bone distraction and cartilage or tendon-ligament fracture.4,33,37
5. Conclusions
The results reveal that hMSCs find both Si and PSi surfaces attractive for developing cell-surface adhesions which are needed in differentiation. The presence of a CM-hMSCs bio-interface improves the differentiation process with respect to a control PSi surface. This was deduced from the following: (i) β-catenin determination showed that cells had increased the characteristic cell–cell adherence of differentiating cells and the fibroblastic morphology involved in cell migration; (ii) the transcription factor STAT-5 was highly expressed and its localization in the cytoplasm indicates a G0 cellular stage; (iii) cells showed a significant increase in nuclear co-localization of the transcription factors Runx2/VDR.These trials provide new evidence of how a CM-hMSCs bio-interface can induce chondrogenesis of bone marrow-derived cells, leading to its consideration in tissue engineering strategies for the development of new scaffolds.
Acknowledgements
We thank Juan García for making the culture medium for hMSCs for so many years. Funding through grant MAT2013-46572-C2-1-R from Ministerio de Economía y Competitividad is greatly appreciated. This work has been partially funded by a UAM-Banco de Santander Interuniversity Cooperation with Asia grant. Hernán Alarcón acknowledges fellowship support from Ministerio de Educación Cultura y Deporte and Fundación Ramón Areces.References
- W. A. Horton, Lancet, 2003, 362, 560–569 CrossRef CAS.
- M. Théry, V. Racine, M. Piel, A. Pépin, A. Dimitrov, Y. Chen, J. B. Sibarita and M. Bornens, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19771–19776 CrossRef PubMed.
- Q. Tseng, E. Duchemin-Pelletier, A. Deshiere, M. Balland, H. Guillou, O. Filhol and M. Théry, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 1506–1511 CrossRef CAS PubMed.
- A. I. Caplan, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 2695–2696 CrossRef CAS PubMed.
- C. R. Rowland, D. P. Lennon, A. I. Caplan and F. Guilak, Biomaterials, 2013, 34, 5802–5812 CrossRef CAS PubMed.
- M. B. Murphy, K. Moncivais and A. I. Caplan, Exp. Mol. Med., 2013, 45, e54 CrossRef PubMed.
- H. Ro, J. Jungha Park, K. Yang, J. Kim, H. G. Yim, G. Jung, H. Lee, S. W. Cho and N. S. Hwang, Cell Tissue Res., 2015 DOI:10.1007/s00441-015-2195-7.
- T. S. de Windt, D. B. Saris, I. C. Slaper-Cortenbach, M. H. van Rijen, D. Gawlitta, L. Creemers, R. A. de Weger, W. Dhert and L. A. Vonk, Tissue Eng. A, 2015 DOI:10.1089/ten.tea.2014.0673.
- A. Díaz-Lantada, H. Alarcón-Iniesta, B. Pareja-Sánchez and J. P. García-Ruiz, Adv. Mater. Sci. Eng., 2014, 2014, 612976 Search PubMed.
- B. Chelli, M. Barbalinardo, F. Valle, P. Greco, E. Bystrenova, M. Bianchi and F. Biscarini, Interface Focus, 2013, 4, 20130041 CrossRef PubMed.
- R. J. Martin-Palma, M. Manso-Silvan and V. Torres-Costa, J. Nanophotonics, 2010, 4, 042502 CrossRef.
- J. Hernández-Montelongo, A. Muñoz-Noval, J. P. García-Ruíz, V. Torres-Costa, R. J. Martín-Palma and M. Manso-Silván, Frontiers in Bioengineering and Biotechnology, 2015, 3, 60 CrossRef PubMed.
- A. Muñoz Noval, V. Sánchez Vaquero, E. Punzón Quijorna, V. Torres Costa, D. Gallach Pérez, L. González Méndez, I. Montero, R. J. Martín Palma, A. Climent Font, J. P. García Ruiz and M. Manso-Silván, J. Biomed. Mater. Res., Part A, 2012, 100A, 1615–1622 CrossRef PubMed.
- M. D. Ynsa, Z. Y. Dang, M. Manso-Silvan, J. Song, S. Azimi, J. F. Wu, H. D. Liang, V. Torres-Costa, E. Punzon-Quijorna, M. B. Breese and J. P. Garcia-Ruiz, Biomed. Microdevices, 2014, 16, 229–236 CrossRef CAS PubMed.
- D. P. Lennon, S. E. Haynesworth, R. G. Young, J. E. Dennis and A. I. Caplan, Exp. Cell Res., 1995, 219, 211–222 CrossRef CAS PubMed.
- S. Ogueta, J. Muñoz, E. Obregon, E. Delgado-Baeza and J. P. García-Ruiz, Mol. Cell. Endocrinol., 2002, 190, 51–63 CrossRef CAS PubMed.
- J. U. Yoo, T. S. Barthel, K. Nishimura, L. Solchaga, A. I. Caplan, V. M. Goldberg and B. Johnstone, J. Bone Jt. Surg., Am. Vol., 1998, 80, 1745–1757 CAS.
- B. Johnstone, T. M. Hering, A. I. Caplan, V. M. Goldberg and J. U. Yoo, Exp. Cell Res., 1998, 238, 265–272 CrossRef CAS PubMed.
- M. F. Pittenger, A. M. Mackay, S. C. Beck, R. K. Jaiswal, R. Douglas, J. D. Mosca, M. A. Moorman, D. W. Simonetti, S. Craig and D. R. Marshak, Science, 1999, 284, 143–147 CrossRef CAS PubMed.
- A. Javed, B. Guo, S. Hiebert, J. Y. Choi, J. Green, S. C. Zhao, M. A. Osborne, S. Stifani, J. L. Stein, J. B. Lian, A. J. van Wijnen and G. S. Stein, J. Cell Sci., 2000, 113, 2221–2231 CAS.
- M. Romero-Prado, C. Blázquez, C. Rodríguez-Navas, J. Munoz, I. Guerrero, E. Delgado-Baeza and J. P. Garcia-Ruiz, J. Cell. Biochem., 2006, 98, 1457–1470 CrossRef CAS PubMed.
- S. Hirohashi and Y. Kanai, Cancer Sci., 2003, 94, 575–581 CrossRef CAS PubMed.
- K. Belmokhtar, T. Bourguignon, M. E. Worou, G. Khamis, P. Bonnet, J. Domenech and V. Eder, Stem Cell Rev. Rep., 2011, 7, 847–859 CrossRef CAS PubMed.
- J. Staerk and S. N. Constantinescu, JAK-STAT, 2012, 1, 184–190 CrossRef PubMed.
- J. Zheng, F. Fang, X. Zeng, T. R. Medler, A. A. Fiorillo and C. V. Clevenger, Mol. Endocrinol., 2011, 25, 2054–2064 CrossRef CAS PubMed.
- A. Villagra, F. Cruzat, L. Carvallo, R. Paredes, J. Olate, A. J. van Wijnen, G. S. Stein, J. B. Lian, J. L. Stein, A. N. Imbalzano and M. Montecino, J. Biol. Chem., 2006, 281, 22695–22706 CrossRef CAS PubMed.
- L. Carvallo, B. Henríquez, R. Paredes, J. Olate, S. Onate, A. J. van Wijnen, J. B. Lian, G. S. Stein, J. L. Stein and M. Montecino, J. Cell. Physiol., 2008, 214, 740–749 CrossRef CAS PubMed.
- S. Marcellini, C. Bruna, J. P. Henríquez, M. Albistur, A. E. Reyes, E. H. Barriga, B. Henríquez and M. Montecino, BMC Evol. Biol., 2010, 10, 78 CrossRef PubMed.
- G. Karsenty and M. Ferron, Nature, 2012, 481, 314–320 CrossRef CAS PubMed.
- J. Wei, J. Shimazu, M. P. Makinistoglu, A. Maurizi, D. Kajimura, H. Zong, T. Takarada, T. Lezaki, J. E. Pessin, E. Hinoi and G. Karsenty, Cell, 2015, 161, 1576–1591 CrossRef CAS PubMed.
- J. Wei and G. Karsenty, Curr. Osteoporosis Rep., 2015, 13, 180–185 CrossRef PubMed.
- X. Yao, R. Peng and J. Ding, Adv. Mater., 2013, 25, 5257–5286 CrossRef CAS PubMed.
- P. A. Netti, Interface Focus, 2013, 4, 20130065 CrossRef.
- S. K. Zaidi, D. W. Young, A. Javed, J. Pratap, M. Montecino, A. van Wijnen, J. B. Lian, J. L. Stein and G. S. Stein, Nat. Rev. Cancer, 2007, 7, 454–463 CrossRef CAS PubMed.
- M. Wuelling and A. Vortkamp, Pediatr. Nephrol., 2010, 25, 625–631 CrossRef PubMed.
- J. K. Laba, A. Steen and L. M. Veenhoff, Curr. Opin. Cell Biol., 2014, 28, 36–45 CrossRef CAS PubMed.
- A. I. Caplan and J. E. Dennis, J. Cell. Biochem., 2006, 98, 1076–1084 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |