Phenylalanine ammonia-lyase, a key component used for phenylpropanoids production by metabolic engineering
Abstract
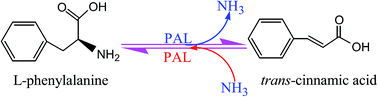
Phenylalanine ammonia-lyase (PAL, EC 4.3.1.24) catalyzes the deamination of phenylalanine to cinnamate and ammonia, the first step of the phenylpropanoid pathway. PALs are ubiquitous in plants and also commonly found in fungi, but have not yet been detected in animals. Typically, PAL is encoded by a small multigene family and the presence of PAL isoforms is a common observation. PAL belongs to the 3,5-dihydro-5-methylidene-4H-imidazol-4-one-containing ammonia-lyase family and has been shown to exist as a tetramer. Both the forward and reverse reactions catalyzed by PALs were of great interest and have potential industrial and medical applications. This review, therefore, covers the recent developments related to the PAL gene distribution, phenylalanine ammonia-lyase gene family, structure and function study of PALs, as well as several potential applications of PALs. As a key gateway enzyme linking the phenylpropanoid secondary pathway to primary metabolism, PALs were extensively applied in heterologous hosts to produce phenylpropanoids. The review thereby highlights the synthetic potentials of PALs as a key component used in metabolic engineering and synthetic biology. Moreover, the other potential PAL applications, like enzyme replacement therapy of phenylketonuria, as a therapeutic enzyme in cancer treatment and microbial production of L-phenylalanine are also discussed in detail. Together these results provide a synopsis of a more global view of potential applications of PALs than previously available.

 Please wait while we load your content...
Please wait while we load your content...