Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biosynthesis and characterization of novel poly(3-hydroxybutyrate-co-3-hydroxy-2-methylbutyrate): thermal behavior associated with α-carbon methylation
Yoriko Watanabe†
*a,
Koya Ishizukaa,
Sho Furutatea,
Hideki Abeab and
Takeharu Tsuge*a
aDepartment of Innovative and Engineered Materials, Tokyo Institute of Technology, 4259 Nagatsuta, Midori-ku, Yokohama 226-8502, Japan. E-mail: watanabe.yoriko@iri-tokyo.jp; tsuge.t.aa@m.titech.ac.jp; Fax: +81-45-924-5426; Tel: +81-45-924-5420
bBioplastic Research Team, RIKEN Biomass Engineering Program, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan
First published on 29th June 2015
Abstract
3-Hydroxy-2-methylbutyrate (3H2MB) has been identified as a minor component of polyhydroxyalkanoates (PHAs) synthesized by bacteria living in activated sludge. In this study, we found that PHA synthase derived from Aeromonas caviae (PhaCAc) polymerizes 3H2MB. By expressing PhaCAc in recombinant Escherichia coli LS5218 and growing cells in the presence of tiglic acid, a PHA copolymer [P(3HB-co-3H2MB)], mainly consisting of 3-hydroxybutyrate (3HB) and up to 37 mol% 3H2MB, was obtained. Analysis of the thermal properties of this novel copolymer indicates that incorporation of 3H2MB into P(3HB) sequence reduced the glass transition temperature (Tg), melting temperature (Tm), and melting enthalpy (ΔHm). The cold crystallization temperature (Tcc) was also lowered by incorporating 7 or 23 mol% 3H2MB, in contrast to the findings for other PHA copolymers. This result suggests that P(3HB-co-3H2MB) copolymers are easier to crystallize than P(3HB) and other PHAs. Thus, 3H2MB provides promising new opportunities to generate 3HB-based polymers with novel thermal properties.
1. Introduction
Polyhydroxyalkanoates (PHAs) are aliphatic polyesters synthesized by diverse bacteria.1–4 Bacteria produce and accumulate PHAs to store energy and carbon when some nutrients such as nitrogen and phosphorus are limited while carbon is sufficient.1 PHAs are of great interest to industry because of their potential as biodegradable and biocompatible thermoplastics.More than 150 different hydroxyalkanoates (HAs) have been identified as building blocks of bacterial PHAs.4 The most typical bacterial PHA is poly[(R)-3-hydroxybutyrate], P(3HB), which is highly crystalline and is brittle and poorly elastic. Less crystalline PHAs are preferred for practical applications such as films and fibers.2 To reduce crystallinity, 3-hydroxyalkanoates (3HAs) with large side chains are copolymerized with 3HB to produce PHA copolymers such as P(3HB-co-3-hydroxyvalerate) [P(3HB-co-3HV)], P(3HB-co-3-hydroxyhexanoate) [P(3HB-co-3HHx)], P(3HB-co-3-hydroxy-4-methylvalerate) [P(3HB-co-3H4MV)], and P(3HB-co-medium-chain-length-3HA) [P(3HB-co-mcl-3HA)]. Currently, these 3HB-based copolymers are considered more preferred than P(3HB).3–7
Many studies have shown that heterotrophic bacteria living in activated sludge accumulate considerable quantities of PHAs. Such PHAs are composed of 3HB and 3HV, as well as derivatives methylated at the α-carbon, namely, 3-hydroxy-2-methylbutyrate (3H2MB) and 3-hydroxy-2-methylvalerate (3H2MV).8–11 PHAs containing such α-carbon methylated units are potentially novel bio-based materials, although PHAs synthesized in sludge bacteria have similar thermal properties as other 3HB-based copolymers.10 This is because ethyl groups in 3HV and 3H2MV units shield the effects of α-carbon methylation. To study the impact of α-carbon methylation on the properties of PHA, P(3HB-co-3H2MB) with controlled monomer composition must be synthesized. To date, P(3HB-co-3H2MB) has never been specifically prepared and characterized.12 Additionally, the bacterial species and the enzyme capable of incorporating 3H2MB into PHA have not been identified.13
PHA synthase derived from Aeromonas caviae (PhaCAc) is an attractive enzyme for PHA synthesis because of relatively wide range of substrate specificity.14 Thus, the aforementioned copolymers such as P(3HB-co-3HHx) and P(3HB-co-3H4MV), which are difficult to be synthesized by other PHA synthases, have been synthesized by PhaCAc.5,7 Additionally, amino acid substitution further broadens the substrate specificity of PhaCAc.15 We have addressed the possibility of a novel PHA synthesis by using PhaCAc and its mutants.
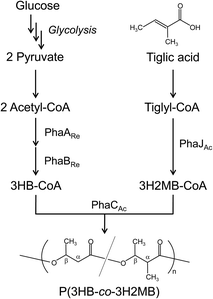
In this study, we found that PhaCAc polymerizes 3H2MB together with 3HB, 3HV, and 3HHx. This finding prompted us to investigate biosynthesis and characterization of a novel P(3HB-co-3H2MB). By expressing PhaCAc in recombinant Escherichia coli LS5218 and growing cells in the presence of tiglic acid, P(3HB-co-3H2MB) with up to 37 mol% 3H2MB was obtained. The copolymers were characterized by gel permeation chromatography (GPC), nuclear magnetic resonance (NMR), and differential scanning calorimetry (DSC), and the effects of α-carbon methylation were investigated. It was revealed that P(3HB-co-3H2MB) has different thermal properties from other 3HB-based copolymers. To our knowledge, this is the first report characterizing P(3HB-co-3H2MB).
2. Experimental section
2.1. Bacterial strain and plasmids
E. coli LS5218 (ref. 16) was used as the host for PHA accumulation. The plasmid pBBR1phaPCJAcABRe NSDG, a variant containing N149S and D171G point mutations in PhaCAc, was constructed by inserting a 0.6-kb PstI-ScaI fragment of pBBREE32d13dPB NSDG17 into the same sites in pBBR1phaPCJAcABRe.18 pBBR1phaPCJAc, which is phaABRe-deficient, and pBBR1phaPCJAc NSDG, which combines phaABRe deficiency with N149S and D171G, were constructed by digesting pBBR1phaPCJAcABRe with FseI, and then treating the resulting 10-kb fragment with Mighty Cloning Reagent Set Blunt End (Takara Bio Inc., Ohtsu, Japan) for blunting, phosphorylation, and ligation. pBBR1phaP(D4N)CJAc, a variant that contains the D4N point mutation in PhaPAc and is phaABRe-deficient,19 and pBBR1phaP(D4N)CJAc NSDG, which combines phaABRe deficiency with all three point mutations, were constructed in the same manner using pBBR1phaP(D4N)CJAcABRe D171L.172.2. PHA synthesis and purification
PHA synthesis was carried out by two-step cultivation. Recombinant E. coli LS5218 was first grown for 4 h at 37 °C with reciprocal shaking (130 strokes per min) in 500 mL flasks containing 100 mL lysogeny broth (LB) media. LB was prepared by dissolving 10 g Bacto-tryptone, 5 g Bacto-yeast extract, and 10 g NaCl in 1 L deionized water. Cells were then harvested by centrifugation at 5960 × g for 10 min at room temperature, washed with sterile deionized water to remove residual media, and resuspended in 1 mL sterile deionized water. The suspension was then inoculated into 100 mL nitrogen-limited M9 media supplemented with glucose (10 g L−1). Nitrogen-limited M9 media was prepared by dissolving the following per liter of distilled water: 17.1 g Na2HPO4·12H2O, 3 g KH2PO4, 0.5 g NH4Cl, 0.5 g NaCl, 2 mL 1 M MgSO4, and 0.1 mL 1 M CaCl2. The pH was adjusted to 7.0. Cultures were then grown at 37 °C for 72 h in a reciprocal shaker (130 strokes per min), and supplemented every 24 h with 0.5 or 1.0 g L−1 trans-2-methylbut-2-enoic acid (tiglic acid), which had been previously converted to its sodium salt. Kanamycin was added to all media at 50 mg L−1 for plasmid maintenance. Polymers accumulated in cells were extracted with chloroform for 72 h at room temperature and purified in two rounds of precipitation with methanol and hexane.2.3. PHA analysis
The composition of isolated PHAs was determined by NMR spectroscopy. Polymers were dissolved in CDCl3 to a final concentration of 20 mg mL−1 and subjected to both 1H- and 13C-NMR using a JEOL LA500 spectrometer. 1H-NMR data were collected at 23 °C with a 7.2 ms pulse width, 90° pulse angle, 5 s pulse repetition, 6000 Hz spectra width, and 16K data points. 13C-NMR data were collected at 25 °C with a 6.1 ms pulse width, 90° pulse angle, 5 s pulse repetition, 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Hz spectra width, and 33K data points. Tetramethylsilane (Me4Si) was used as internal chemical shift standard.
000 Hz spectra width, and 33K data points. Tetramethylsilane (Me4Si) was used as internal chemical shift standard.
The amount and composition of PHA accumulated in lyophilized cells were determined by GC and GC-MS after methanolysis in the presence of 15% sulfuric acid.20 GC was carried out in a Shimadzu GC-14B system with a non-polar capillary column (InertCap1, 30 m × 0.25 mm, GL Sciences Inc., Tokyo, Japan) and a flame ionization detector, while GC-MS was performed using GCMS-QC 2010 (Shimadzu, Kyoto, Japan).
Molecular weight was measured by GPC at 40 °C using a Shimadzu 10A GPC system, with a 10A refractive index detector and two Shodex K806M columns. Chloroform was used as eluent at a flow rate of 0.8 mL min−1, and samples were applied at 3.0 mg mL−1. Polystyrene standards with low polydispersity were used to obtain a calibration curve.
To analyze PHA by DSC, 4–5 mg of PHA film was encapsulated in aluminum pans and heated under nitrogen with a Perkin-Elmer Pyris 1 DSC (Perkin-Elmer, Waltham, MA, USA) from −50 to 200 °C with a heating rate of 20 °C min−1 (first heating scan). The melt samples were then maintained at 200 °C for 1 min and followed by rapid quenching at −50 °C. They were heated from −50 to 200 °C at a heating rate of 20 °C min−1 (second heating scan). Prior to analysis, films were aged over 3 weeks at room temperature to reach equilibrium crystallinity.
2.4. Hydratase activity
Cell extracts were prepared from E. coli BL21(DE3) harboring the expression plasmid pETNB3,14 which contains A. caviae R-hydratase PhaJAc, or pETJ1,21 which contains Pseudomonas aeruginosa R-hydratase PhaJ1Pa. Briefly, recombinant cells were inoculated in 100 mL LB media with 100 mg L−1 ampicillin and grown at 37 °C in a reciprocal shaker (130 strokes per min). After 3 h, cultures were induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside. Cells were harvested at 3 h after induction and lysed by sonication. Lysates were cleared by centrifugation for 30 min at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 × g and 4 °C. The soluble fraction was loaded directly onto a HiLoad Q-Sepharose HP 16/10 column (Amersham Biosciences, Piscataway, NJ, USA) pre-equilibrated with 20 mM Tris–HCl, pH 7.5. Bound proteins were eluted over a 250 mL linear gradient from 0 to 1.0 M NaCl, at a flow rate of 2.5 mL min−1. The purified enzyme was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein concentration was determined with Quant-iT™ Protein Assay Kits (Invitrogen, Carlsbad, CA, USA) using bovine serum albumin as the standard.
600 × g and 4 °C. The soluble fraction was loaded directly onto a HiLoad Q-Sepharose HP 16/10 column (Amersham Biosciences, Piscataway, NJ, USA) pre-equilibrated with 20 mM Tris–HCl, pH 7.5. Bound proteins were eluted over a 250 mL linear gradient from 0 to 1.0 M NaCl, at a flow rate of 2.5 mL min−1. The purified enzyme was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein concentration was determined with Quant-iT™ Protein Assay Kits (Invitrogen, Carlsbad, CA, USA) using bovine serum albumin as the standard.
Enoyl-CoA hydratase activity was measured by hydration of trans-2-enoyl-CoA substrates. Diluted PhaJ (5 μL) was added to 895 μL of 50 mM Tris–HCl, pH 8.0, containing 25 μM trans-2-enoyl-CoA, and the decrease in absorbance (ε) at 263 nm was measured at 30 °C using a quartz cuvette with 1.0 cm light path. The ε263 of the enoyl-thioester bond is 6.7 × 103 M−1 cm−1. The substrates crotonyl-CoA and tiglyl-CoA were synthesized, based on a mixed-anhydride method, from crotonic acid (Tokyo Kasei, Tokyo, Japan) and tiglic acid (Kanto Chemical, Tokyo, Japan), respectively, with a lithium salt of CoA.22 Products were purified on a Sep-Pak C18 column (Waters, Milford, MA, USA).23
3. Results and discussion
3.1. Synthesis of PHA copolymers containing 3H2MB
PHA copolymers containing 3H2MB have been produced in active sludge, using organic acids as the carbon source,8–11 although the bacterial species and PHA synthase responsible have not been identified.13 In this study, biosynthesis of 3H2MB-containing copolymers from tiglic acid was achieved by expressing the enzyme PhaCAc (from A. caviae) in E. coli LS5218, which is a mutant strain that utilizes fatty acids more efficiently than other E. coli strains.16The yield and composition of PHAs obtained in a two-step cultivation scheme are summarized in Table 1. Cells expressing phaPCJAc (pBBR1phaPCJAc) accumulated 1 wt% PHA with 3.1 mol% of 3H2MB. This finding prompted us to develop a strain that synthesizes P(3HB-co-3H2MB) using PhaCAc. Subsequently, we discovered that the NSDG mutant of PhaCAc, which contains the N149S and D171G point mutations,15 showed increase in PHA accumulation and 3H2MB incorporation to 2.4 wt% and 14.8 mol%, respectively. This result suggests the NSDG mutant polymerizes 3H2MB more efficiently than the wild-type.
| Exogenous genes | Glucose (g L−1) | Tiglic acid (g L−1) | Dry cell weight (g L−1) | PHA content (wt%) | PHA compositionb (mol%) | Sample no. | |||
|---|---|---|---|---|---|---|---|---|---|
| 3HB | 3H2MB | 3HV | 3HHx | ||||||
| a Cells harboring pBBR1-MCS2 derivatives were cultivated in two-step cultures, in which cells were first cultured in LB media and subsequently transferred to nitrogen-limited M9 media with or without 10 g L−1 glucose, and supplemented with 0.5 or 1.0 g L−1 tiglic acid every 24 h for 72 h. Results are mean ± standard deviation of three separate experiments. n.d.: not detectable.b Determined by GC.c N149S and D171G point mutations in PhaCAc.d D4N point mutation in PhaPAc. | |||||||||
| phaPAc, phaCAc, phaJAc | 10 | 1 g L−1 × 3 | 2.2 ± 0.1 | 1.0 ± 0.1 | 96.9 | 3.1 | n.d. | n.d. | 1 |
| phaPAc, phaCAc(NSDG)c, phaJAc | 10 | 1 g L−1 × 3 | 2.0 ± 0.1 | 2.4 ± 0.4 | 83.5 | 14.8 | 0.3 | 1.4 | 2 |
| phaPAc(D4N)d, phaCAc, phaJAc | 10 | 1 g L−1 × 3 | 2.1 ± 0.1 | 6.0 ± 0.8 | 77.3 | 19.0 | 2.6 | 1.1 | 3 |
| phaPAc(D4N), phaCAc(NSDG), phaJAc | 10 | 1 g L−1 × 3 | 2.2 ± 0.1 | 8.8 ± 0.5 | 63.6 | 33.9 | 1.0 | 1.5 | 4 |
| phaPAc(D4N), phaCAc(NSDG), phaJAc | 10 | 0.5 g L−1 × 3 | 2.1 ± 0.1 | 11 ± 0.5 | 76.0 | 22.0 | 0.6 | 1.4 | 5 |
| phaPAc(D4N), phaCAc(NSDG), phaJAc | — | 1 g L−1 × 3 | 0.9 ± 0.1 | n.d. | — | — | — | — | 6 |
| phaPAc(D4N), phaCAc(NSDG), phaJAc, phaARe, phaBRe | 10 | 1 g L−1 × 3 | 2.5 ± 0.2 | 34 ± 0.4 | 88.5 | 10.9 | 0.3 | 0.3 | 7 |
3.2. Enhanced PHA accumulation by PhaPAc mutation
During the course of PhaCAc studies, we unexpectedly found that one plasmid increased PHA accumulation in E. coli. DNA sequencing revealed that this plasmid contained the point mutation D4N in the PHA granule-associated protein (PhaPAc).18 This mutation enhances the expression of the entire phaPCJ operon.19 Here, we examined the effect of PhaPAc D4N on 3H2MB polymerization, and found that PHA accumulation and 3H2MB incorporation increased to 6.0 wt% and 19.0 mol%, respectively (Table 1). However, 3HV and 3HHx were also incorporated more efficiently, so that they comprised 2.6 mol% and 1.1 mol%, respectively, of PHA.Furthermore, we observed a synergistic effect when PhaPAc D4N was combined with PhaCAc NSDG. As a result, PHA with 33.9 mol% 3H2MB was accumulated to 8.8 wt%, without a significant increase in 3HV and 3HHx content. Taken together, these results suggest that PhaPAc D4N enhances not only PHA accumulation but also 3H2MB incorporation.
3.3. Compositional control via feeding or PhaAB expression
In an attempt to further enhance 3H2MB incorporation, glucose was completely replaced with tiglic acid. However, the cells did not accumulate PHA (Table 1). In addition, we attempted to manipulate PHA accumulation and 3H2MB incorporation by varying the amount of tiglic acid used. Thus, when the concentration of tiglic acid was reduced to 0.5 g L−1 every 24 h, 22.0 mol% incorporation of 3H2MB was achieved, with relatively high PHA accumulation of 11 wt%. Indeed, by controlling the amount of tiglic acid in the media, the fraction of 3H2MB incorporated could be varied between 0 and 33.9 mol%.Finally, we co-expressed enzymes that synthesize 3HB (PhaABRe) with PhaCAc NSDG and PhaPAc D4N (Table 1). The resulting copolymer contained 10.9 mol% 3H2MB with little 3HV and 3HHx (0.3 mol% each). PHA accumulation was further increased to 34 wt%, the highest value reached in this study.
3.4. NMR analysis of P(3HB-co-3H2MB)
PHAs were extracted from samples 4, 5, and 7 (characteristics described in Table 1) and analyzed by NMR. The 1H-NMR spectrum of P(3HB-co-3H2MB) at 500 MHz is shown in Fig. 1. Peaks at 5.3 ppm (b), 2.4–2.6 ppm (a), and 1.3 ppm (c) were assigned to the methine, methylene, and methyl proton resonances, respectively, of 3HB. Peaks at 5.1 (e), 2.7 (d), 1.2 (f), and 1.1 ppm (g) were assigned to be methine, methylene, and methyl proton resonances, respectively, at the β position and methyl proton resonances at the α position of 3H2MB. In addition, peaks at 1.7 (h) and 0.9 ppm (i) were assigned to methylene and methyl proton resonances of 3HA side chains, respectively. In sample 4, the intensity ratio of peaks (d), (e), (f), and (g) was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, which is in good agreement with the ratio estimated from the structure of 3H2MB. The composition of this copolymer, as calculated from the intensity ratio of peaks (c), (f) and (i), was 60 mol% 3HB, 37 mol% 3H2MB, and 3 mol% 3HA. The composition of other samples were determined in the same manner, and summarized in Table 2.
3, which is in good agreement with the ratio estimated from the structure of 3H2MB. The composition of this copolymer, as calculated from the intensity ratio of peaks (c), (f) and (i), was 60 mol% 3HB, 37 mol% 3H2MB, and 3 mol% 3HA. The composition of other samples were determined in the same manner, and summarized in Table 2.
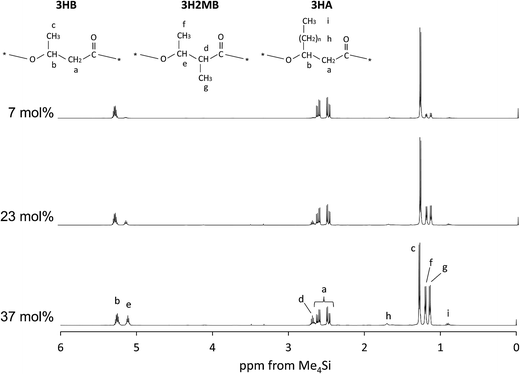 | ||
| Fig. 1 The 500 MHz 1H-NMR spectrum of P(3HB-co-3H2MB) containing 7 mol% (sample 7), 23 mol% (sample 5), and 37 mol% 3H2MB (sample 4). | ||
| Sample no. or name | PHA compositiona (mol%) | Thermal propertiesc | Molecular weight | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 3HB | 3H2MB | 3HAb | Tg (°C) | Tcc (°C) | Tm (°C) | ΔHm (J g−1) | Mn (×103) | Mw/Mn | |
| a Composition determined by 1H-NMR.b 3HV + 3HHx.c Tg, glass-transition temperature (2nd heating); Tcc, cold crystallization temperature (2nd heating); Tm, melting temperature (1st heating); ΔHm, enthalpy of fusion (1st heating).d P(3HB) homopolymer obtained from R. eutropha H16. | |||||||||
| P(3HB)d | 100 | 0 | 0 | 4 | 58 | 178 | 97 | 228 | 2.3 |
| 7 | 92 | 7 | 1 | 1 | 46 | 146, 157 | 57 | 1300 | 3.1 |
| 5 | 75 | 23 | 2 | −1 | 39 | 140, 150 | 35 | 96 | 6.4 |
| 4 | 60 | 37 | 3 | −1 | 57 | 131, 138 | 17 | 90 | 4.5 |
Additional structural evidence for incorporation of 3H2MB was obtained in 13C-NMR spectra at 125 MHz. In Fig. 2, the strong peaks 1–4 were assigned to carbon resonances of 3HB, while some of the weaker peaks 5–9 could be assigned to carbon resonances of 3H2MB.8,9
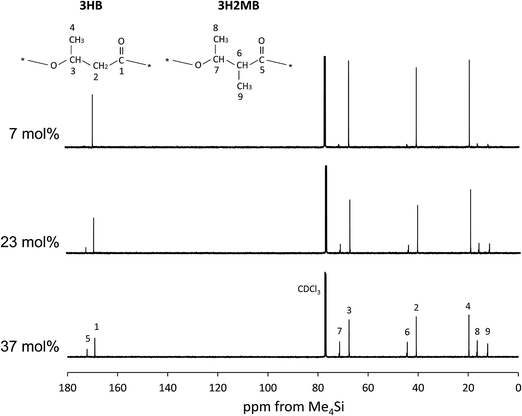 | ||
| Fig. 2 The 125 MHz 13C-NMR spectrum of P(3HB-co-3H2MB) containing 7 mol% (sample 7), 23 mol% (sample 5), and 37 mol% 3H2MB (sample 4). | ||
3.5. Molecular weight of P(3HB-co-3H2MB)
GPC analysis was carried out to determine the molecular weight of P(3HB-co-3H2MB) samples. Molecular weights are listed in Table 2. Samples 4 and 5 exhibited a low Mn of (90–96) × 103 but high Mw/Mn of 4.5–6.4. In contrast, the Mn of sample 7 was as high as 1300 × 103, with Mw/Mn of 3.1, probably because PhaABRe was expressed in these cells. The enzymes would have increased the pool of 3HB monomers available, and thus could potentially enhance elongation of polymer chains. Such high molecular weights were also observed for the strains co-expressing phaPCJAc and phaABRe in a previous study.193.6. Thermal properties of PHA containing 3H2MB
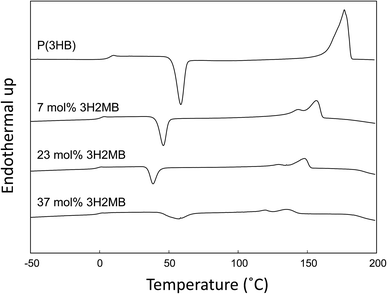
DSC thermograms of copolymers are shown in Fig. 3, and thermal properties are summarized in Table 2. As the 3H2MB fraction increased from 0 to 37 mol%, the glass transition temperature (Tg), melting temperature (Tm) and melting enthalpy (ΔHm) decreased from 4 °C to −1 °C, 178 °C to 131 °C, and 97 to 17 J g−1, respectively.Generally, Tg, Tm, and ΔHm of 3HB-based copolymers such as P(3HB-co-3HV) and P(3HB-co-3HHx) decrease in proportion to the fraction of 3HV or 3HHx incorporated. For P(3HB-co-3HV) with 25 mol% 3HV, the Tg, Tm, and ΔHm were approximately 3 °C, 120 °C and 70 J g−1, respectively.24 The high ΔHm value of P(3HB-co-3HV) is attributed to the occurrence of cocrystallization of 3HB and 3HV units. As shown in Table 2, the Tg, Tm, and ΔHm of P(3HB-co-23 mol% 3H2MB) were −1 °C, 131 °C and 17 J g−1, respectively. Although 3H2MB is a structural isomer of 3HV, the thermal properties of P(3HB-co-3H2MB) are quite different from those of P(3HB-co-3HV); incorporation of 3H2MB results in a copolymer with higher Tm but lower ΔHm.
3.7. Crystallization behavior of P(3HB-co-3H2MB)
As shown in Fig. 3 and Table 2, the cold crystallization temperature (Tcc) of P(3HB-co-3H2MB), as observed on DSC second heating scan, decreased from 58 °C to 39 °C with incorporation of 23 mol% 3H2MB. However, PHA with 37 mol% 3H2MB had Tcc of 57 °C, which might be due to increased incorporation of 3HA, at 3 mol%.It is known that ΔT = Tcc − Tg is an indication of the crystallization behavior of polymers;25 the smaller the ΔT, the easier the polymer tends to crystallize. The ΔT for P(3HB) and P(3HB-co-23 mol% 3H2MB) are 54 °C and 40 °C, respectively, suggesting that the latter is easier to crystallize than the former. On the other hand, low-molecular weight polymers tend to easily crystallize; however, P(3HB-co-7 mol% 3H2MB) also showed a low ΔT even though its molecular weight was very high. Thus, α-methylation significantly influenced the crystallization propensity of P(3HB). Finally, we compared ΔT of various 3HB-based copolymers (Fig. 4), and found that the crystallization propensity of P(3HB-co-3H2MB) did not follow the same general trend as those of other 3HB-based copolymers,5,7,26 regardless of whether or not the cocrystallization occurred. These results suggest that 3H2MB does not inhibit P(3HB) crystallization, thereby conferring novel thermal properties to 3HB-based polymers.
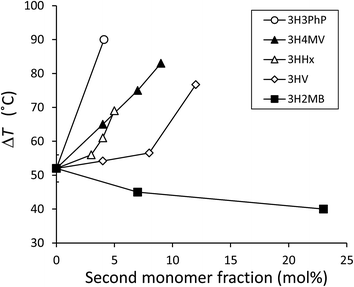 | ||
| Fig. 4 Relationship between ΔT (ΔT = Tcc − Tg) of 3HB-based copolymers measured at 20 °C min−1 of heating rate and the molar fraction of the copolymerizing unit up to 23 mol%. ΔT for P(3HB-co-3HV) was measured using commercially available samples. ΔT for other copolymers was obtained from past reports.5,7,26 From these reports, the average ΔT for P(3HB) was calculated to be 52 ± 4 °C and was plotted in this figure as the y-intercept. 3HV, 3-hydroxyvalerate; 3H4MV, 3-hydroxy-4-methylvalerate; 3HHx, 3-hydroxyhexanoate; 3H3PhP, 3-hydroxy-3-phenylpropionate. | ||
3.8. Hydratase activity of PhaJAc
To measure the ability of PhaJAc to synthesize 3H2MB, its hydratase activity was assayed using crotonyl-CoA and tiglyl-CoA as substrates and R-hydratase (PhaJ1Pa) from P. aeruginosa as ref. 23. The results are listed in Table 3. PhaJAc hydrated tiglyl-CoA and crotonyl-CoA at 7450 and 0.18 U mg−1, respectively. The ratio between these two activities was 2.4 × 10−5. In comparison, PhaJ1Pa also showed hydration activities with a comparable activity ratio of 3.7 × 10−5. Thus, this PhaJAc has the ability to generate 3H2MB-CoA from tiglyl-CoA, presumably according to the pathway shown in Fig. 5. We also examined other PhaJs, including PhaJ4Pa from P. aeruginosa23 and PhaJAc mutants.27 However, significant enhanced activity was not detected in any of these enzymes (data not shown).| Enzyme (origin) | Hydratase activity (U mg−1) | Activity ratio (B/A) × 10−5 | |
|---|---|---|---|
| Crotonyl-CoA (A) | Tiglyl-CoA (B) | ||
| a All values are mean ± standard deviation of triplicate assays. | |||
| PhaJAc (A. caviae) | 7450 ± 990 | 0.18 ± 0.01 | 2.4 |
| PhaJ1Pa (P. aeruginosa) | 4200 ± 940 | 0.16 ± 0.01 | 3.7 |
4. Conclusion
In this study, P(3HB-co-3H2MB) synthesis was demonstrated in E. coli LS5218 expressing PhaCAc and using tiglic acid as the 3H2MB precursor. 3H2MB incorporation up to 37 mol% was achieved, while incorporation of other 3HA was successfully inhibited to 3 mol%. Analysis of thermal properties revealed that, unlike other PHA copolymers, P(3HB-co-3H2MB) showed relatively low ΔHm with relatively high Tm. Notably, researches to improve PHA has focused on decreasing ΔHm by copolymerization, usually resulting in a severe decrease in Tm. Thus, this study provides alternative and new approaches to improve PHA. Furthermore, P(3HB-co-3H2MB) shows a small temperature difference between Tg and Tcc, suggesting that it crystallizes more easily than P(3HB) and other 3HB-based copolymers, presumably due to α-carbon methylation. Slow crystallization of P(3HB) and PHA copolymers is a significant drawback for the use of these materials in melt processing; P(3HB-co-3H2MB) may overcome this defect. In summary, the thermal properties of P(3HB-co-3H2MB) are of great interest, as 3H2MB appears to generate 3HB-based polymers that are different from others. Further study is now ongoing to determine configuration of the α-methyl group in 3H2MB and comonomer compositional distributions of P(3HB-co-3H2MB), which may explain the unconventional properties of this copolymer.Acknowledgements
We thank Y. Nakamura (Tokyo Institute of Technology) for NMR analysis. This work was partially supported by funding from JST, CREST.Notes and references
- D. Jendrossek and D. Pfeiffer, Environ. Microbiol., 2014, 16, 2357–2373 CrossRef CAS PubMed.
- K. Sudesh, H. Abe and Y. Doi, Prog. Polym. Sci., 2000, 25, 1503–1555 CrossRef CAS.
- R. W. Lenz and R. H. Marchessault, Biomacromolecules, 2005, 6, 1–8 CrossRef CAS PubMed.
- B. H. A. Rehm, Curr. Issues Mol. Biol., 2007, 9, 41–62 CAS.
- T. Tsuge, Y. Saito, Y. Kikkawa, H. Hiraishi and Y. Doi, Macromol. Biosci., 2004, 4, 238–242 CrossRef CAS PubMed.
- I. Noda, P. R. Green, M. M. Satkowski and L. A. Schechtman, Biomacromolecules, 2005, 6, 580–586 CrossRef CAS PubMed.
- N. Tanadchangsaeng, A. Kitagawa, T. Yamamoto, H. Abe and T. Tsuge, Biomacromolecules, 2009, 10, 2866–2874 CrossRef CAS PubMed.
- H. Satoh, T. Mino and T. Matsuo, Int. J. Biol. Macromol., 1999, 25, 105–109 CrossRef CAS.
- Y. Inoue, F. Sano, K. Nakamura, N. Yoshie, Y. Saito, H. Satoh, T. Mino, T. Matsuo and Y. Doi, Polym. Int., 1996, 39, 183–189 CrossRef CAS.
- S. Bengtsson, A. R. Pisco, P. Johansson, P. C. Lemos and M. A. M. Reis, J. Biotechnol., 2010, 147, 172–179 CrossRef CAS PubMed.
- H. Satoh, T. Mino and T. Matsuo, Water Sci. Technol., 1992, 26, 933–942 CAS.
- B. Füchtenbusch, D. Fabritius and A. Steinbüchel, FEMS Microbiol. Lett., 1996, 138, 153–160 CrossRef.
- A. Michinaka, J. Arou, M. Onuki, H. Satoh and T. Mino, Biotechnol. Bioeng., 2007, 96, 871–880 CrossRef CAS PubMed.
- T. Fukui, N. Shiomi and Y. Doi, J. Bacteriol., 1998, 180, 667–673 CAS.
- T. Tsuge, S. Watanabe, D. Shimada, H. Abe, Y. Doi and S. Taguchi, FEMS Microbiol. Lett., 2007, 277, 217–222 CrossRef CAS PubMed.
- S. K. Spratt, C. L. Ginsburgh and W. D. Nunn, J. Bacteriol., 1981, 146, 1166–1169 CAS.
- Y. Watanabe, Y. Ichinomiya, D. Shimada, A. Saika, H. Abe, S. Taguchi and T. Tsuge, J. Biosci. Bioeng., 2012, 113, 286–292 CrossRef CAS PubMed.
- A. Saika, Y. Watanabe, K. Sudesh and T. Tsuge, J. Biosci. Bioeng., 2014, 117, 670–675 CrossRef CAS PubMed.
- K. Ushimaru, Y. Watanabe, A. Hiroe and T. Tsuge, J. Gen. Appl. Microbiol., 2015, 61, 63–66 CrossRef PubMed.
- M. Kato, H. J. Bao, C. K. Kang, T. Fukui and Y. Doi, Appl. Microbiol. Biotechnol., 1996, 45, 363–370 CrossRef CAS.
- T. Tsuge, K. Taguchi, S. Taguchi and Y. Doi, Int. J. Biol. Macromol., 2003, 31, 195–205 CrossRef CAS.
- J. C. Fong and H. Schulz, Methods Enzymol., 1981, 71, 390–398 CAS.
- H. E. Valentin and A. Steinbüchel, Appl. Microbiol. Biotechnol., 1994, 40, 699–709 CrossRef CAS.
- Y. Wang, S. Yamada, N. Asakawa, T. Yamane, N. Yoshie and Y. Inoue, Biomacromolecules, 2001, 2, 1315–1323 CrossRef CAS PubMed.
- X. Q. Liu, M. X. Wang, Z. C. Li and F. M. Li, Macromol. Chem. Phys., 1999, 200, 468–473 CrossRef CAS.
- S. Mizuno, S. Katsumata, A. Hiroe and T. Tsuge, Polym. Degrad. Stab., 2014, 109, 379–384 CrossRef CAS PubMed.
- T. Tsuge, T. Hisano, S. Taguchi and Y. Doi, Appl. Environ. Microbiol., 2003, 69, 4830–4836 CrossRef CAS.
Footnote |
| † Present address: Tokyo Metropolitan Industrial Technology Research Institute, 2-4-10 Aomi, Koto-ku, Tokyo 135-0064, Japan. |
| This journal is © The Royal Society of Chemistry 2015 |