Multi-functional fluorescent carbon dots with antibacterial and gene delivery properties†
Qingqing Doua,
Xiaotian Fanga,
Shan Jianga,
Pei Lin Cheea,
Tung-Chun Lee d and
Xian Jun Loh*abc
d and
Xian Jun Loh*abc
aInstitute of Materials Research and Engineering, A*STAR, (Agency for Science, Technology and Research), 3 Research Link, Singapore 117602, Singapore. E-mail: lohxj@imre.a-star.edu.sg; XianJun_Loh@scholars.a-star.edu.sg; Tel: +65-65131612
bDepartment of Materials Science and Engineering, National University of Singapore, Singapore 117574, Singapore
cSingapore Eye Research Institute, 20 College Road, Singapore 169856, Singapore
dInstitute for Materials Discovery, University College London, Kathleen Lonsdale Building, Gower Place, WC1E 6BT London, UK
First published on 13th May 2015
Abstract
Glucose is abundant in nature and can be found in various sources. In this study, we developed multifunctional carbon dots (CDs) with glucose and poly(ethyleneimine) (PEI), which were further quaternized using a facile approach. The CDs are designed to possess both anti-bacteria and gene delivery capabilities. The inherent property was characterized with TEM, NMR, FTIR and fluorescent spectroscopy. Antibacterial activity was evaluated with Broth minimum inhibitory concentration (MIC) assay on both Gram-positive and Gram-negative bacteria. The CDs showed excellent inhibitation to both bacteria. The expression of CDs condensed plasmid DNA in HEK 293T cells was investigated with luciferase expression assay. Gene transfection capability of the quaternized CDs was found to be up to 104 times efficient than naked DNA delivery.
Antibacterial materials are of great importance in wound disinfection and implantable medical devices, and are closely related to our daily life. The materials widely investigated for antibacterial purpose includes antibiotics,1 noble metal nanoparticles2,3 and quaternary ammonium compounds.4 The above mentioned materials give rise to problems such as antibiotic resistance, high cost and non-environmental friendly synthesis methods. Since the serendipitous discovery of carbon dots (CDs) in 2004,5 concerted efforts have been devoted to synthetic and property studies on them as they show superior fluorescent property and bio-friendly. Carbon dots has been synthesized with citric acid and ethylenediamine,6 phenylboronic acid,7 gum Arabic,8 orange juice,9 and commercial food.10 Glucose, abundant material in nature, has also been reported as the precursor for the synthesis of carbon dots in the presence of ammonium hydroxide,11 monopotassium phosphate12 and other alkali or acid.13 These methods involve harsh basic or acidic conditions. Furthermore, carbon dots which are made from carbon sources such as glucose typically promote bacterial growth, promoting contamination and hindering their applications in cell culture assays. Here we report a facile method to synthesize anti-microbial carbon dots with glucose, where we use poly(ethyleneimine) (PEI) to achieve synthesis-passivation integration in one step. PEI has been widely used in cosmetics, pharmaceuticals, water treatment, detergents and adhesives.14 Notably, PEI has shown great permeabilization properties with Gram-negative bacteria.15,16 As reported by Kawabata and Nishiguchi,17 quaternized polymer exhibit strong antibacterial activity against Gram-positive bacteria as well. Here, we aim to inhibit both Gram-negative and positive bacteria by introducing PEI and quaternizing the amine groups in the carbon dots with benzyl bromide. We studied the effect of PEI structure on carbon dots as passivation agent by using branched or linear PEI in the synthesis. Later, Broth microdilution minimum inhibitory concentration test was performed to evaluate antibacterial effect of the quaternized CDs on both Gram-negative (Escherichia coli) and Gram-positive (Staphylococcus aureus) bacteria. PEI is also well reported as a gene transfection agent. In order to evaluate its utility as a gene transfection agent, luciferase assay was performed in HEK 293T cells to evaluate gene transfection capability of the quaternized CDs.
The CD synthesis was carried out by the hydrothermal method and was carried on in a Teflon-lined, stainless steel autoclave. In a typical synthesis, 15 mL aqueous solution with 1.0 g of PEI and 0.3 g of glucose was autoclaved at 150 °C for 12 hours. The effect of the structure of the capping agent on surface passivation was investigated by altering branched PEI (25 kDa) or linear PEI (10 kDa) in the reaction, of which products denote as CDb and CDl respectively.
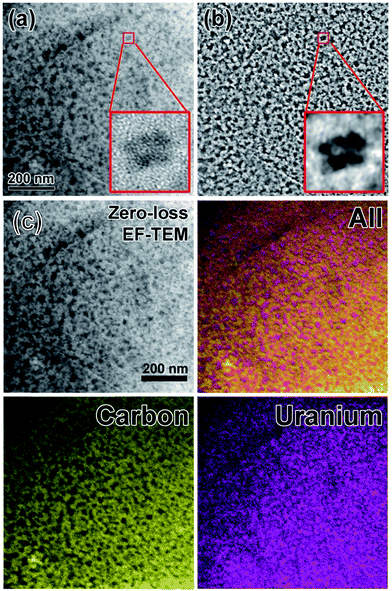
The size distribution of the as-synthesized CDs was measured by ZetaPlus Brookhaven Instruments with dynamic light scattering (DLS). The results showed that the size of CDs was neither affected by the structure nor the amount of PEI used for passivation (Fig. S1†). On the other hand, energy-filtered TEM images obtained from Zeiss Omega 912 at 120 kV accelerating voltage showed that the mean size of CDs is 3.5 nm with a standard deviation 0.9 nm, counted from more than 200 particles. The size is consistent with DLS data. The TEM grid used here was firstly glow discharged for 3 min followed by twice washing with DI water, and subsequently stained with 0.5% uranyl acetate (UA) for 1.5 min for an enhanced contrast. The preferential staining of the carbon dots (over the carbon membrane of the grid) is due to the high affinity of uranyl ions to sialic acid carboxyl groups derived from modified glucose in CDs. Energy filter was inserted to select only the zero-loss peak (i.e. elastically scattered electrons) to further enhance the contrast of the bright field TEM image. A band pass image filter was applied to filter out features <5 pixels and >50 pixels on the zero-loss bright field TEM image to get better resolved images (Fig. 1b). Since the sample was negatively stained by UA, it may be debatable that the zoom-in image in Fig. 1a is the carbon dots. To clarify this doubt, three-window technique was applied on the zero-loss energy-filtered TEM images for elemental mapping (Fig. 1c). False colors were assigned to carbon and Uranium for easy recognition. The results reconciled the band pass filtered TEM image in Fig. 1b.
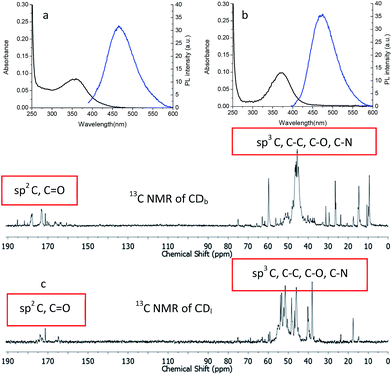
1H NMR was used to characterize the chemical structure of CDs. The chemical shift between δ = 2.2–2.9 ppm (Fig. 2) was attributed to the CH2N bonds18,19 in the PEI-passivated carbon dots, where the N is derived from PEI. In contrast, the control 1H NMR spectrum of glucose did not show signal in this range. Moreover, there was a small peak at δ = 8.3 ppm observed only in both branched and linear PEI-passivated carbon dots, but not in the PEIs and glucose. This was indicative of a presence of (CO)–N–H, and the presence of a hydrogen attached to an amide nitrogen,20 which suggests the formation of a bond between PEI and the glucose core in the carbon dots. Furthermore, the washing steps with chloroform did not appear to affect the PEI on the carbon dots surface.
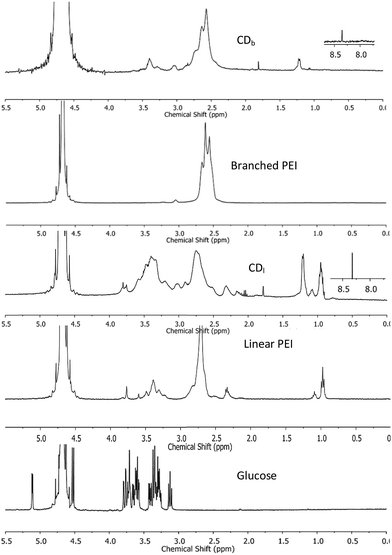 | ||
| Fig. 2 1H NMR spectra of CDs passivated with branched PEI (CDb) and linear PEI (CDl), control spectra obtained from branched and linear PEI, and glucose. | ||
The optical property of the CDs was characterized with UV-Vis absorption spectrum on a Shimadzu UV-3150 spectrometer and photoluminescence (PL) emission spectrum with Shimadzu RF-5301 PC spectrofluorophotometer. Branched PEI capped CDb showed maximum absorbance at 360 nm, while the linear PEI capped CDl maximum absorbance was 370 nm (Fig. 3). The shoulder absorption peak at 300–400 nm was attributed to n–π* transitions of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O.21
O.21
When excited the CDs at 360 nm, the emission centered at 466 nm and 473 nm for CDb and CDl, respectively. The little difference should arise from the effect of PEI structure to the inter-layer interaction between adjunct sp2 islands, which is the emission center of CDs.22,23 Moreover, the chemical shifts δ = 160–184 ppm (Fig. 3c) in 13C NMR corresponds to sp2 carbons in carbonyl (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O).24,25 Aliphatic sp3 carbon atoms were also observed, evidenced by chemical shifts between δ = 30–45 ppm. As the linear structure fits in more easily between the sp2 islands compared to branched PEI, linear PEI disrupts the inter-layer π–π connections between the sp2 islands. Thus, the CDs passivated with linear PEI showed relatively low fluorescence.
O).24,25 Aliphatic sp3 carbon atoms were also observed, evidenced by chemical shifts between δ = 30–45 ppm. As the linear structure fits in more easily between the sp2 islands compared to branched PEI, linear PEI disrupts the inter-layer π–π connections between the sp2 islands. Thus, the CDs passivated with linear PEI showed relatively low fluorescence.
Excitation wavelength dependent emission of the CDs was observed. The emission peak center exhibited red shift when using a longer wavelength excitation, which is in agreement with other researchers' findings.26–28 Branched PEI passivated CDs (CDb) showed higher capacity of color tuning (from 463 to 525 nm) than the CDs modified with linear PEI (CDl), but the emission intensity were gradually decreased.
Besides the sp2 island mechanism of the CDs emission, another mechanism hypothesis was that radiative recombination of the excited electrons from the n–π* transitions of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in multiple surface emissive trap states lead to CD emision.28,29 It can be seen from the 13C NMR (Fig. 3c), carbon in sp3 state was also present in the CDs. The presence of multiple electronic structures could form a series of energy transitions between bandgaps and a pathway to radiative recombination. More possible recombination renders CDs responsive to different wavelength excitation, and each would show a unique emission. The strongest transition was found between 330–360 nm as shown in UV-Vis spectrum in Fig. 4a and b, and the emission diminished as the excitation wavelength was shifted beyond this range. This explains the excitation dependant emission phenomenon in CDs.
O in multiple surface emissive trap states lead to CD emision.28,29 It can be seen from the 13C NMR (Fig. 3c), carbon in sp3 state was also present in the CDs. The presence of multiple electronic structures could form a series of energy transitions between bandgaps and a pathway to radiative recombination. More possible recombination renders CDs responsive to different wavelength excitation, and each would show a unique emission. The strongest transition was found between 330–360 nm as shown in UV-Vis spectrum in Fig. 4a and b, and the emission diminished as the excitation wavelength was shifted beyond this range. This explains the excitation dependant emission phenomenon in CDs.
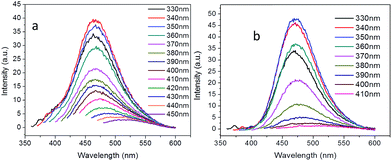 | ||
| Fig. 4 Excitation-dependent PL emission of the carbon dots passivated with (a) branched PEI (CDb) and (b) linear PEI (CDl). | ||
With quinine sulfate (QS) in 0.10 M H2SO4 as the reference, absolute quantum yield of CDs was measured with eqn (1).
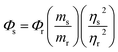 | (1) |
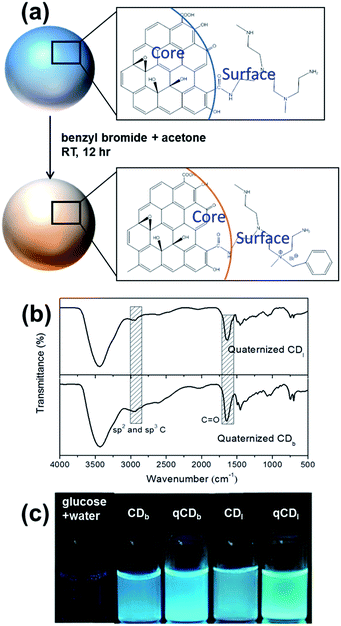
Benzyl bromide was quaternized to the PEIs on carbon dots in a simple procedure at room temperature. Typically, CDs and benzyl bromide in the ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (w/v) were dispersed in deionized water. After adding two times the volume of acetone, the mixture was stirred for 24 hours at room temperature. The illustration of this reaction is shown in Fig. 5a. The FTIR spectra (Fig. 5b) showed an absorption peak around 1660–1700 cm−1, which covers C
5 (w/v) were dispersed in deionized water. After adding two times the volume of acetone, the mixture was stirred for 24 hours at room temperature. The illustration of this reaction is shown in Fig. 5a. The FTIR spectra (Fig. 5b) showed an absorption peak around 1660–1700 cm−1, which covers C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching in conjugation with phenyl group and conjugation with double bond (1680–1700 cm−1). The characteristic peak at 2900–3000 cm−1 was assigned to sp2 and sp3 C–H stretching. The fluorescence and quantum yield of the carbon dots was not affected by quaternization as shown in the optical images (Fig. 5c).
O stretching in conjugation with phenyl group and conjugation with double bond (1680–1700 cm−1). The characteristic peak at 2900–3000 cm−1 was assigned to sp2 and sp3 C–H stretching. The fluorescence and quantum yield of the carbon dots was not affected by quaternization as shown in the optical images (Fig. 5c).
The anti-microbial ability of the quaternized CDs was accessed by the Broth minimum inhibitory concentration (MIC). MIC is a standard test of antimicrobial activities of compounds that inhibit the growth of bacteria after 24 hours of incubation with bacteria.32 A Gram-positive bacterium Staphylococcus aureus (S. aureus) and a Gram-negative bacterium Escherichia coli (E. coli) were used for MIC testing. After incubation with a serial of CDs dilution, the percentage of survived bacteria in the media was quantified by the absorbance at 600 nm. The growth of the bacteria can be evaluated as eqn (2).
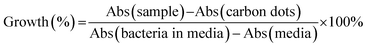 | (2) |
The findings in the MIC results showed that quaternary CDs efficiently inhibited both Gram-positive and Gram-negative bacteria growth. It was previously reported that PEI has shown great permeabilization properties with Gram-negative bacteria,15,16 and linear PEIs induced depolarization of S. aureus membrane.33 Furthermore quaternary linear PEI passivated CDs (qCDb) showed improved inhibition to S. aureus compared to qCDb. Similarly, quaternary linear PEI passivated CDs also better inhibited growth of Gram-negative E. coli. The antibacterial property of the quaternized CDs is due to qCDs be able to absorb onto the cytoplasmic membrane and causing its disruption.34,35
As PEI is an often used polymeric gene transfection agent, gene transfection capability of the quaternized CDs was evaluated with luciferase expression assay in HEK 293T cells. Plasmid pRL-CMV, which encodes Renilla luciferase, was used as the reporter gene for the gene expression.36–41 The plasmid DNA (pDNA) was condensed by quaternized CDs via electrostatic attraction before being transfected in HEK 293T cells. After CDs were taken up by cells, the condensed pDNA were released into cells and the luciferase expressed in cells. Based on the luminescence intensity of luciferase, the gene delivery efficiency can be evaluated. CDs/pDNA complexes, prepared at CDs/pDNA weight ratios of 0.75, 1.5, 3, 4.5, 7.5 and 15, containing 1.0 μg of pDNA were examined in the gene transfection experiment. The results showed that the branched PEI modified CDs exhibited higher gene transfection efficiency than linear PEI (Fig. 6). The best gene expression was found in the sample with CDs/pDNA complexes at ratio of 7.5. At lower weight ratio, pDNA cannot be condensed efficiently by the cationic CDs, and the resultant loose CDs/pDNA complex cannot enter the cell easily. At higher CDs/pDNA ratios, the cytotoxicity of an increasing amount of free CDs may result in a reduction in the transfection efficiency. In contrast, naked pDNA only showed very low transfection (<104). Fig. 6 showed that qCDs improved the gene transfection capability up to 104 fold for qCDb and about 103 folds for qCDl, respectively (Table 1).
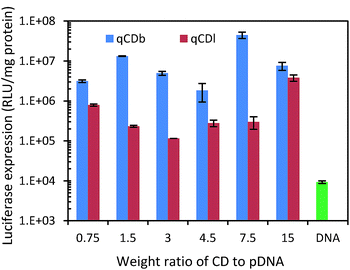 | ||
| Fig. 6 Luciferase expression test of quaternized carbon dots and pDNA only to evaluate gene delivery capability. | ||
| Sample ID | MIC (μg mL−1) | |
|---|---|---|
| Gram-positive bacteria | Gram-negative bacteria | |
| S. aureus | E. coli | |
| qCDb | 32 | 64 |
| qCDl | 16 | 16 |
Conclusions
In conclusion, we have developed multifunctional carbon dots (CDs) with antibacterial and gene transfection properties. The CDs are derived from natural abundant material – glucose and PEI, and eventually quaternized with benzyl bromide. Unlike other methods for surface passivation of CDs, the synthesis method we developed to obtain the material is facile and simple, which avoided using harsh acid or base. With this approach, the fluorescent property of CDs, such as quantum yield and emission wavelength, can be tuned by changing the structure of PEI, which affects the inter-layer sp2 islands interaction in CDs. The quarternization with benzyl bromide bestows the CDs not only with antibacterial but also gene delivery functions. The quaternized carbon dots exhibited promising antibacterial activity against both Gram-negative (Escherichia coli) and Gram-positive (Staphylococcus aureus) bacteria. We have also explored the potential of these CDs for gene transfection together with synchronized imaging of the fate of CDs in a hygienic manner without worrying about bacterial contaminations.Notes and references
- M. Wenzel, M. Patra, C. H. R. Senges, I. Ott, J. J. Stepanek, A. Pinto, P. Prochnow, C. Vuong, S. Langklotz and N. Metzler-Nolte, ACS Chem. Biol., 2013, 8, 1442–1450 CrossRef CAS PubMed.
- S. Chernousova and M. Epple, Angew. Chem., Int. Ed., 2013, 52, 1636–1653 CrossRef CAS PubMed.
- R. R. Arvizo, S. Bhattacharyya, R. A. Kudgus, K. Giri, R. Bhattacharya and P. Mukherjee, Chem. Soc. Rev., 2012, 41, 2943–2970 RSC.
- S. Buffet-Bataillon, P. Tattevin, M. Bonnaure-Mallet and A. Jolivet-Gougeon, Int. J. Antimicrob. Agents, 2012, 39, 381–389 CrossRef CAS PubMed.
- X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS PubMed.
- S. Zhu, Q. Meng, L. Wang, J. Zhang, Y. Song, H. Jin, K. Zhang, H. Sun, H. Wang and B. Yang, Angew. Chem., Int. Ed., 2013, 52, 3953–3957 CrossRef CAS PubMed.
- P. Shen and Y. Xia, Anal. Chem., 2014, 86, 5323–5329 CrossRef CAS PubMed.
- M. Thakur, S. Pandey, A. Mewada, V. Patil, M. Khade, E. Goshi and M. Sharon, J. Drug Delivery, 2014, 2014, 9 Search PubMed.
- S. Sahu, B. Behera, T. K. Maiti and S. Mohapatra, Chem. Commun., 2012, 48, 8835–8837 RSC.
- M. P. Sk, A. Jaiswal, A. Paul, S. S. Ghosh and A. Chattopadhyay, Sci. Rep., 2012, 2, 383 Search PubMed.
- Z. Ma, H. Ming, H. Huang, Y. Liu and Z. Kang, New J. Chem., 2012, 36, 861–864 RSC.
- Z.-C. Yang, M. Wang, A. M. Yong, S. Y. Wong, X.-H. Zhang, H. Tan, A. Y. Chang, X. Li and J. Wang, Chem. Commun., 2011, 47, 11615–11617 RSC.
- H. Li, X. He, Y. Liu, H. Huang, S. Lian, S.-T. Lee and Z. Kang, Carbon, 2011, 49, 605–609 CrossRef CAS PubMed.
- M. Noh, Y. Mok, D. Nakayama, S. Jang, S. Lee, T. Kim and Y. Lee, Polymer, 2013, 54, 5338–5344 CrossRef CAS PubMed.
- I. M. Helander, H.-L. Alakomi, K. Latva-Kala and P. Koski, Microbiology, 1997, 143, 3193–3199 CrossRef CAS PubMed.
- I. M. Helander, K. Latva-Kala and K. Lounatmaa, Microbiology, 1998, 144, 385–390 CrossRef CAS PubMed.
- N. Kawabata and M. Nishiguchi, Appl. Environ. Microbiol., 1988, 54, 2532–2535 CAS.
- Y.-B. Lim, C. E. Mays, Y. Kim, W. B. Titlow and C. Ryou, Biomaterials, 2010, 31, 2025–2033 CrossRef CAS PubMed.
- F. A. Carey and R. M. Giuliano, Organic Chemistry, McGraw-Hill, 2011 Search PubMed.
- D. Pavia, G. Lampman, G. Kriz and J. Vyvyan, Introduction to Spectroscopy, Cengage Learning, 2014 Search PubMed.
- Z. Gan, S. Xiong, X. Wu, T. Xu, X. Zhu, X. Gan, J. Guo, J. Shen, L. Sun and P. K. Chu, Adv. Opt. Mater., 2013, 1, 926–932 CrossRef PubMed.
- G. Eda, Y.-Y. Lin, C. Mattevi, H. Yamaguchi, H.-A. Chen, I. S. Chen, C.-W. Chen and M. Chhowalla, Adv. Mater., 2010, 22, 505–509 CrossRef CAS PubMed.
- H. Li, Z. Kang, Y. Liu and S.-T. Lee, J. Mater. Chem., 2012, 22, 24230–24253 RSC.
- Y. Liu, Y. Tian and W. Yang, RSC Adv., 2014, 4, 58758–58761 RSC.
- H. E. Gottlieb, V. Kotlyar and A. Nudelman, J. Org. Chem., 1997, 62, 7512–7515 CrossRef CAS.
- G. Wang, X. Pan, J. N. Kumar and Y. Liu, Carbon, 2015, 83, 180–182 CrossRef CAS PubMed.
- X. Li, S. Zhang, S. A. Kulinich, Y. Liu and H. Zeng, Engineering surface states of carbon dots to achieve controllable luminescence for solid-luminescent composites and sensitive Be2+ detection, Sci. Rep., 2014, 4, 4976 CAS.
- Y.-P. Sun, B. Zhou, Y. Lin, W. Wang, K. A. S. Fernando, P. Pathak, M. J. Meziani, B. A. Harruff, X. Wang, H. Wang, P. G. Luo, H. Yang, M. E. Kose, B. Chen, L. M. Veca and S.-Y. Xie, J. Am. Chem. Soc., 2006, 128, 7756–7757 CrossRef CAS PubMed.
- S. Y. Lim, W. Shen and Z. Gao, Chem. Soc. Rev., 2015, 44, 362–381 RSC.
- Q. Hu, M. C. Paau, Y. Zhang, X. Gong, L. Zhang, D. Lu, Y. Liu, Q. Liu, J. Yao and M. M. F. Choi, RSC Adv., 2014, 4, 18065–18073 RSC.
- S. Chandra, P. Patra, S. H. Pathan, S. Roy, S. Mitra, A. Layek, R. Bhar, P. Pramanik and A. Goswami, J. Mater. Chem. B, 2013, 1, 2375–2382 RSC.
- J. M. Andrews, J. Antimicrob. Chemother., 2001, 48, 5–16 CrossRef CAS PubMed.
- K. Gibney, I. Sovadinova, A. I. Lopez, M. Urban, Z. Ridgway, G. A. Caputo and K. Kuroda, Macromol. Biosci., 2012, 12, 1279–1289 CrossRef CAS PubMed.
- C. Wiegand, M. Bauer, U.-C. Hipler and D. Fischer, Int. J. Pharm., 2013, 456, 165–174 CrossRef CAS PubMed.
- E.-R. Kenawy, S. D. Worley and R. Broughton, Biomacromolecules, 2007, 8, 1359–1384 CrossRef CAS PubMed.
- Y. Zeng, R. Yi and B. R. Cullen, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9779–9784 CrossRef CAS PubMed.
- X. J. Loh, S. J. Ong, Y. T. Tung and H. T. Choo, Mater. Sci. Eng., C, 2013, 33(8), 4545–4550 CrossRef CAS PubMed.
- X. J. Loh, S. J. Ong, Y.T. Tung and H. T. Choo, Macromol. Biosci., 2013, 13(8), 1092–1099 CrossRef CAS PubMed.
- X. J. Loh, J. Appl. Polym. Sci., 2013, 127(2), 992–1000 CrossRef CAS PubMed.
- X. J. Loh, S. J. Ong, Y. T. Tung and H. T. Choo, Polym. Chem., 2013, 4(8), 2564–2574 RSC.
- X. J. Loh, Z.-X. Zhang and K. Y. Mya, et al., J. Mater. Chem., 2010, 20(47), 10634–10642 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra07968c |
| This journal is © The Royal Society of Chemistry 2015 |