Isolation of doxorubicin from a bacterial culture using immobilised metal ion affinity chromatography†
Abstract
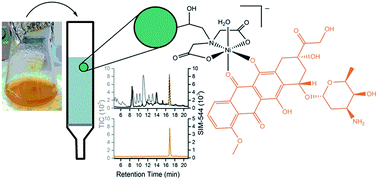
The restricted supply of the anticancer agent doxorubicin (DXR) prompted the development of a simple method to isolate DXR from a Streptomyces peucetius var. caesius liquid culture that could facilitate its local production. The metal-coordinating ability of DXR enabled its purification using immobilised metal ion affinity chromatography (IMAC). The entirety of a DXR standard (94 nmol) bound to 1 mL of Ni(II)-charged iminodiacetic acid (IDA) IMAC resin at pH 7.5 and was eluted as a free ligand at pH 5.5. The DXR binding capacity of the Ni(II)-charged IDA resin was about 4.7 μmol mL−1. An aliquot of the crude S. peucetius liquid culture containing native DXR (∼2 nmol) was retained on 1 mL of the Ni(II)-charged IDA resin in 38%. The reduced binding capacity was likely due to competing ligands in the bacteriological medium. Re-application of a sub-sample of the unbound DXR on a second column gave 81% DXR retention. The utility of IMAC to purify DXR on an analytical scale warrants further evaluation of industrial-scale IMAC processing as a potential route to this essential medicine.

 Please wait while we load your content...
Please wait while we load your content...