Dual role of pinostrobin-a flavonoid nutraceutical as an efflux pump inhibitor and antibiofilm agent to mitigate food borne pathogens†
Abstract
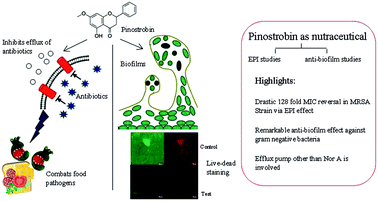
Nutraceutical pinostrobin displayed synergy with ciprofloxacin against diverse bacteria causing drastic 128 fold reduction in ciprofloxacin MIC in MRSA and significant antibiofilm effect against E. coli and P. aeruginosa at subinhibitory concentrations. Studies with NorA mutants of S.aureus revealed efflux inhibition of pinostrobin is mediated by efflux pumps other than NorA.


 Please wait while we load your content...
Please wait while we load your content...