Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Tuning neuron adhesion and neurite guiding using functionalized AuNPs and backfill chemistry
Pinggui
Li
ac,
Kyrylo
Greben
ac,
Roger
Wördenweber
ac,
Ulrich
Simon
bc,
Andreas
Offenhäusser
ac and
Dirk
Mayer
*ac
aPeter Grünberg Institute (PGI-8) and Institute of Complex Systems (ICS-8), Forschungszentrum Jülich GmbH, 52428 Jülich, Germany. E-mail: dirk.mayer@fz-juelich.de
bInstitute of Inorganic Chemistry, RWTH Aachen University, 52074 Aachen, Germany
cJARA – Fundamentals of Future Information Technology, Germany
First published on 23rd April 2015
Abstract
The adhesion of neurons depends on the interplay between attractive as well as repellant cues in the cell membrane and adhesion ligands in their cellular environment. In this study, an easy and versatile strategy is presented to control the density of cell binding sites embedded in a cell repulsive environment attached to a solid surface. Gold nanoparticles modified by positively charged aminoalkyl thiols are used as artificial neuron adhesion ligands. The density of the nanoparticles and their environment is varied by applying either no backfill, poly(ethylene glycol)-silane, or octyltrichlorosilane backfill. By this means the chemical composition of both cell attractive adhesion ligands and surrounding repellant cues is tuned on the nanometer scale. Primary rat cortical neurons are cultured on these particle modified surfaces. The viability and neuritogenesis of neurons is investigated as a function of particle density and background composition. A strong dependence of neuron viability on both averaged particle density and backfill composition is found in particular for intermediate particle packing. At high particle densities, the kind of backfill does not affect the cell viability but influences the development of neurites. This knowledge is used to enhance the guiding efficiency of neuron adhesion to more than 90% on nanopatterned surfaces.
1. Introduction
The control of cell adhesion is of increasing interest for the investigation of fundamental aspects of cell development such as cell differentiation, cell growth, tissue development and regeneration. Furthermore, neuronal adhesion to artificial substrates is a key element of biotechnological and biomedical neuroscience.1–3 Cellular bioelectronic research including, for instance, the development of neuronal implants, neuronal prostheses, and cell-based biosensors often relies on the firm attachment of neurons with solid-state devices to achieve precise interfacing with the cells.4–6 A close and stable contact between neuron and device surface is important to ensure a small seal resistance and an optimal communication between them.7 Mainly three cues of solid surfaces are influencing adhesion of neurons: the chemical composition of the surface,12–14 compliance,8–11,15–17 and topographical contrast.1,18–20 As chemical adhesion cues mainly cell adhesive molecules (CAMs),21,22 neurotrophic factors,23 ligands of specific guidance receptors,24,25 extra cellular matrix proteins such as laminin and fibronectin or selected oligopeptides of those like RGD and IKVAV are used.26–30 Also corollaries of the specific surface composition such as the wettability31 and surface charges32 are influencing cellular behavior on biomaterial surfaces. It has been recognized that in particular positively charged domains of peptides assist the adhesion of neurons.33 This finding was supported by the observation that also positive charged amino groups associated with surface bound synthetic molecules can promote the adhesion and growth of neurons.34–37In vivo studies showed that positive charges aid the regeneration of nerves.38 Many cell membranes possess a high number of glycosylated proteins which are contributing to the formation of a negative net charge at the extracellular part of the membrane. The intercellular recognition and adhesion is regulated by these negatively charged carbohydrates. For neurons, the polysialylation of neural cell adhesion molecule (NCAM) facilitates cell migration and plasticity by regulating the repulsion between adjacent cells.39 The electrostatic interactions between the negatively charged glycocalyx and cationic adhesion cues has been used to immobilize and pattern cells on solid surfaces.40With the advances in nanofabrication, it has become feasible to define the chemical composition of solid surfaces on the nanoscale and to study their effect on cellular behavior.41–43 It has been demonstrated that nanoscale objects are able to influence cellular properties and processes such as cell shape, cell adhesion, alignment, differentiation and others.44–46 The reason for the strong impact of nanoobjects on cellular features is that many cell functions are controlled by proteins which possess nanoscale dimensions. Therefore, it has been proposed that nanoscience may help achieve a greater precision and parallelism in electrical and chemical sensing of cell/neuron activity.47
Protein patterns as small as 75 nm were generated by microcontact printing and used to control the adhesion of primary neurons and directed outgrowth of neurites.48 J. H. Huang et al. arranged gold nanoparticles with varied interparticle spacing as carriers of single peptide guidance factors in patterned arrays on substrates by diblock copolymer micelle nanolithography.49 Their studies indicate that the spatial organization of the extracellular environment is important in regulating cell–extra cellular matrix (ECM) interactions and thus cell adhesion.21,26,41,50–53 In addition to the chemical influence of nanoparticles on cells, they allow to obtain additional analytical information about the investigated system due to their optical properties.54,55 We recently developed a simple and efficient strategy to deposit randomly close-packed AuNPs on a solid surface by electrostatic interactions.56 In this article we investigate the variation of the surfaces composition at nanometer scale by changing both the density of the particles and the chemical composition of the particle surrounding. The AuNPs were functionalized by 11-amino-1-undecanethiol (AUT), a molecule that possesses a positive charge under neuron culture conditions. The amino-functionalized gold nanoparticle (AF-AuNP) can be considered as cationic anchor spots for glycosylated plasma membrane proteins similar to widely used cationic polyamino acids like polylysines, however, with strongly confined size and control over density and local distribution. Samples homogeneously modified with poly-D-lysine (PDL) are used as control to evaluate the maturation of the neurons. We investigate the influence of the densities of surface bound particle on the viability of primary rat cortical neurons. In addition, we modified the surrounding background of the AF-AuNP to vary the chemical contrast between the particle and their environment. This implicates that we control the lateral distribution of cell attractive and cell aversive materials at the nanometer scale and thereby tune the cell adhesive properties of the solid surface on micron and millimeter scale. We investigate the influence of three common background materials: negatively charged SiO2, 2-[methoxy(polyethyleneoxy)6-9-propyl] trichlorosilane (PEG silane), and octyltrichlorosilane (OTS). The bare, hydrophilic SiO2 was supposed to have cell repellent properties since it possesses an excess of negative charges at culture conditions and should therefore repulse the glycocalyx of the neurons. PEG silane was chosen as backfill molecule because of its excellent protein repulsive property to minimize unspecific interaction between proteins and inorganic surfaces.49,57,58 In addition, we used OTS as backfill material since it was demonstrated that hydrophobic surfaces inhibit cell adhesion.59 We found that rat cortical neurons can adhere to AF-AuNPs functionalized surfaces and the cell viability at high particle densities was even better than for PDL control samples. Furthermore, we observed that not only ligand density is critical for neuronal adhesion49 but also the chemical contrast between the AF-AuNPs and environment play a vital role in attachment of neurons on solid surfaces and the outgrowth of neurites. We used the gained knowledge to optimize the guiding efficiency of micro- and nano-patterned particle modified surfaces. The guiding of neuron adhesion and neurite outgrowth is not only required for the construction of defined in vitro neural networks but also for the alignment of neurons with solid-state devices to realize bioelectronic hybrid systems.60–63 For this purpose various patterning techniques been used such as photolithography and microcontact printing.64,65 These patterning techniques have in common that they generate a local pattern with homogenous chemical composition on micrometer scale. This corresponds to a kind of “on” and “off” situation for the adhesion of cell and their processes from the nanometer scale perspective. Based on a large area top down nanoimprint lithography we generated locally defined chemical templates for the bottom up self-assembly of nanoparticle on these template patterns. Thereby, we control the lateral distribution of cell attractive and cell aversive materials at the nanometer scale. By this means we can enhance the chemical contrast between particle free and particle containing areas which results in a guiding efficiency for rat cortical neurons of more than 90%.
2. Results
2.1 Characterization of particle modified surfaces
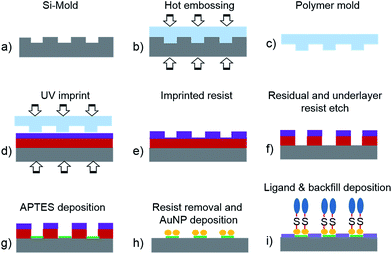
The preparation and characterization of the AuNP modified surfaces has been described in detail in previous publications.56,66 Just briefly, a plasma activated SiO2 surface is modified by aminopropylsilane molecules and citrate stabilized AuNP are immobilized on this surface via electrostatic interactions. Subsequently the particle decorated surface is activated in an oxygen plasma and the particle surrounding SiO2 surface is modified by octyltrichlorosilane (OTS) or 2-[methoxy(polyethyleneoxy)6-9-propyl] trichlorosilane (PEG silane) from vapor phase. Finally, the sample is immersed into an 11-amino-1-undecanethiol solution to modify the surface of gold particles (Scheme 1).In the following we report on the characterization of sample preparation steps that have not been part of previous reports as for instance the backfilling. Therefore, we first of all characterized the wettability of silicon surfaces before and after modifying the surface by a backfill layer. We used sessile drop contact angle measurements to obtain static contact angles. After O2 plasma activation of the bare silicon surface, we obtained a perfectly hydrophilic surface with a contact angle of θ < 10°. After the PEG silane graft, the static contact angle increases however the surface remained hydrophilic with a contact angle of 44° ± 1° similar to previous reports.67,68 At contact angles smaller than 50° ethylene glycol surfaces become adsorption resistant against hydrophilic and hydrophobic peptides.57 For the OTS silanization we observed a change of the surface wettability from hydrophilic to hydrophobic and an increase of the static contact angle to (82° ± 2°).69
Furthermore, we evaluated the successful attachment of the 11-amino-1-undecanethiol to AuNP decorated Si/SiO2 surfaces with and without PEG silane backfill by means of X-ray photoelectron spectroscopy (XPS). In both cases, we prepared samples with a high density of particles of about 500 particles per μm2 (see below). For both surfaces we observed a high content of Si, oxygen, and Au which are originating from the sample substrate and the AuNP core, respectively, Table 1. The signals of these elements decreased after AUT binding due to the formation of a molecular layer on the gold particles attenuating substrate signals. In addition we found nitrogen as well as sulfur which can be assigned to the formed AUT shell on the AuNP. The sulfur and nitrogen signals are relatively weak since the particles decorate just parts of the sample (less than 20%) and the information depth of this technique is much larger than the length of AUT. Furthermore, the S2s signal is attenuated by the alkyl chain capping the thiol group. The fact that the sulfur and nitrogen signal can be observed also after application of a PEG backfill layer to the AF-AuNP free areas of SiO2 shows that the backfill does not impair the formation of particle associated gold thiolate bonds.
| AuNP + O2 plasma (at.%) | AuNP + AUT (at.%) | |
|---|---|---|
| Si | 58 | 44 |
| S | 0 | 1 |
| Au | 21 | 13 |
| N | 0 | 2 |
| C | 21 | 40 |
2.2 Particle density assays
Citrate stabilized AuNP were immobilized from colloidal solutions of different concentration on a self-assembled monolayer (SAM) of aminopropylsilane. With increasing particle concentration, the density of particles on the surface gradually increases as described previously.66 By this simple method we were able to tune the density of particles from 4 ± 2 to 541 ± 20 particles per μm2 with a coverage changing from 0.1 to around 20% by simply diluting the particle solution from 0.0001 to 0.01 wt%, correspondingly. The SEM images reveal a random distribution of the AuNPs on silica surface (Fig. 1) at different concentrations. The particles are well dispersed. Aggregation of two or more particles occur rarely. By comparing the particle density before and after backfill, we found that about 5% of the particles were released from the surface due to the backfill treatment.2.3 Neuronal culture assay
Rat embryonic cortical neurons were cultured on AUT modified AuNPs with different particle densities and different backgrounds to investigate the influence of the particle distance and chemical background on the adhesion and viability of neurons. We used a serum free neuron basal medium for cell culture in the scope of this work. Since we intend to control the adhesion of electrogenic neurons and the outgrowth of their neurites we aim to avoid a domination of the culture by glia cell after long culture times. To identify the cell type, we performed an immunostaining to distinguish neuron from glial (results not shown). We found only very few glia cells on the sample after 9 DIV of all surface treatments tested. The ratio of glia vs. neuron was typically smaller than 0.07 indicating a successful suppression of glia proliferation.The adhered neurons were co-stained with calcein AM (green) and ethidium homodimer (orange) to indicate healthy and dead cells, respectively, after 5 days in vitro (DIV). We observed an enhanced viability of the neurons with increasing particle density for all samples tested for PEG silane backfill, Fig. 1. For low particle density, most cells appear as small red dot which indicates cell death and a detachment of the cell from the sample surface. With higher particle density an increasing number of vital cells stained in green are visible. The samples with the highest particle density provided accordingly the best neuron culture conditions among the tested samples.
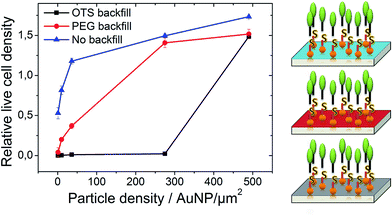
For a qualitative assessment of the neuron viability on the different particle modified surfaces we determined the relative live cell density for all samples and plotted this quantity vs. the particle density. The relative live cell density is calculated by dividing the live cell density of a particle containing sample by the live cell density of the corresponding PDL control sample. From the resulting plot remarkable differences become apparent for samples with different particle environments, Fig. 2. Samples that have been modified with particles but without backfill facilitate neuron adhesion even for the lowest particle density. With increasing number of particles the relative live cell density rises steeply, however, saturates at approx. 1.7. This means that not only the live cell density increases with the particle density but also that the density of vital neurons is higher on surfaces with high particle density than on PDL control samples. The final neuron viability for samples with high particle densities is about three times higher than the cell density without any particle.
After modifying the particle environment of the SiO2 surface by PEG silane backfill, the adhesion of neurons is strongly suppressed for the lowest particle densities (4 particle per μm2), which is in accordance with previous observations.70–72 Only very few cells can survive on PEG backfilled substrate but in an unhealthy state. With increasing particle densities the relative live cell density increases until it saturates. For each specific particle density, the neuron viability was larger for samples without backfill than with PEG backfill. Also the saturation occurs at higher particle densities for backfill modified samples. In the case of a hydrophobic OTS backfill, we found a poor neuron adhesion for the lowest particle density which increases only marginally until the cell viability steeply increased at a particle density of approx. 500 particle per μm2. Remarkably, the final relative neuron density is very similar for all surface modifications independent of the used backfill.
To evaluate the effectiveness of the backfill, we plated neurons on bare, PEG silane, and OTS modified SiO2 surfaces without any AF-AuNP. The bare SiO2 surface does not facilitate a complete suppression of neuron adhesion (Fig. 3 top left). Almost the same relative live cell density was found (0.5) for these samples as for AF-AuNP surfaces with low particle densities 4 particle per μm2. In addition we observed, that the neurons are able to spread from particle containing (500 particle per μm2) to particle free areas of the surface without clear border between these domains (Fig. 3 top right). The number of vital cells is higher on the particle modified regions, however, the neurons manage to mature also on bare SiO2. On the contrary, no vital cells could be observed on bare PEG silane modified SiO2 (Fig. 3 middle left). For samples modified with AF-AuNP and PEG silane backfill we observed a sharp border between particle containing and particle free areas (Fig. 3 middle right). Neither somas are able to adhere nor do neurites manage to grow out from particle containing to particle free areas. OTS modified samples showed a similar behavior (Fig. 3 bottom). Apparently, both PEG silane and OTS efficiently suppress neuron adhesion while SiO2 only reduces partially cell viability although no neuron binding ligands have been applied to the surface. To evaluate the influence of the backfill onto the neural development and neurite outgrowth, we cultured neurons at 9 DIV on AF-AuNP modified surfaces without backfill, with PEG silane, or OTS backfill. All samples possessed a high particle density of approx. 500 particles per μm2. Subsequently, we stained the neurons for axons and dendritic neurites by rabbit anti-MAP2 primary antibodies and mouse anti-Tau1 antibodies, respectively.
The neurons cultured on backfill free particle surfaces show a matured development with soma diameters in the range of 20 to 30 μm (Fig. 4 top). Each neuron possesses in average 6 short dendritic neurites which are typically shorter than 100 μm and stained orange. The axons stained in green have a length of more than several hundred microns. At this stage of development the neuron axons tend to loop back and to form networks of crossing axonal extensions. The morphological appearance of the neurons is similar to those of neurons cultured on PDL control samples or on extra cellular matrix proteins at glia containing co-culture conditions.73 Neurons cultured on surfaces modified by AF-AuNP and PEG silane backfill exhibit a development that is similar to the backfill free AF-AuNP samples, however the number of processes and length of axons are slightly smaller, Table 2. Neurons plated on OTS backfilled samples have a different appearance. Although the number of neurons adhered to the surface as well as the number of neurites are comparable to the other two samples, length of the neuron processes is significantly smaller. An analysis of the branching of neurites revealed that the influence of AF-AuNP on the formation of higher order branches is small.
 | ||
| Fig. 4 MAP-2 antibody staining dendrites (orange) and Tau1 antibody staining axon (green) at 5 DIV: top-without backfill, middle-with PEG backfill, bottom-with OTS backfill (scale bar: 100 μm). | ||
| Substrate | Neurites number | Axon length (μm) |
|---|---|---|
| PDL | 7.2 ± 0.9 | 380 ± 67 |
| Particles without backfill | 6.1 ± 0.8 | 368 ± 69 |
| Particles with PEG backfill | 5.4 ± 0.9 | 302 ± 59 |
| Particles with OTS backfill | 6.2 ± 0.9 | 130 ± 76 |
To further evaluate the adhesion of the cortical neurons on particle modified surfaces we investigated the samples by scanning electron microscopy. The high resolution capabilities of this technique allows a detailed imaging of the spreading of the neurite processes on the surface and simultaneous mapping of particles and neurons. Exemplary SEM images of neurons at 9 DIV cultured on AF-AuNP surfaces are shown in Fig. 5.
The cell somas tightly spread on the particle film with many processes extending to the cell environment. The neurites themselves possesses many sub-micron sized filopodial processes that are contacting the surface of nanoparticles. Cross sections of the sample surface/neuron interface obtained from FIB cutting of fixated cells demonstrate that the particles are not internalized by the neurons and still associated to the sample surface even after long culturing time in vitro. This observation further confirms that the backfill does not impair the binding strength of the particles to the sample surface. The electrostatic interaction between AF-AuNPs and the APTES functionalized SiO2 surface is strong enough to hold the particles on the surface even after cells interact with the particles in a cell culture medium (Fig. 5 bottom).74 Also the top view SEM images do not reveal any evidence of a displacement (particle free areas next to neuron extensions) or accumulation of particles due to mechanical actuations of the cells.
We did not observe any significant difference of the surface/particle/neuron interface for all kinds of backfills investigated by us.
In our previous work we used micro- and nanostructured AF-AuNP patterns for guiding of neuron adhesion. We achieved the guiding of neurons by establishing an adhesion contrast between particle associated positively charged amino groups and the SiO2 substrate with predominant negative surface charges at neuron culture conditions. By this means we achieved a guiding efficiency of about 71%.56 The results described above, however, revealed that the difference of the viability of neurons on particle free SiO2 surfaces is just a factor of three smaller than for surfaces with a high density of particle associated binding ligands. Furthermore, we found that the chemical contrast to AF-AuNP can be enhanced by applying a backfill to particle surrounding areas. Therefore, we evaluated the guiding efficiency for the same AF-AuNP pattern as described previously however this time in combination with an OTS backfill. The particle surface patterns were established on oxidized silicon samples by a process using nanoimprint lithography (NIL), particle immobilization, and backfilling, Scheme 2.
The pattern consisted of 8.2 μm-wide stripes separated by gaps of 11.6 μm. The stripes are sub-structured in micron-sized squares of 10.1 μm × 10.1 μm and line packages. The latter are composed of nano-sized lines of 200 to 400 nm widths and 300 nm gaps in between. The micron-scaled squares should provide nodes for a conformal adhesion of the neurons, whereas the features consisting of nano-scaled lines were adapted to the size of neurites and their extensions. After plating cortical neurons onto the patterned surface, the cells preferably adhere onto those areas that are decorated by high density AF-AuNP. At 3 DIV, a distinct majority of the neurons possess a bidirectional neurite outgrowth with processes longer than 100 μm. A branching of neuritis orthogonal to the particle stripes and crossing of the particle free areas occurs relatively seldom although the distance between the stripes is just 11.6 μm. The cells are well aligned to the particle pattern, Fig. 6. The guiding efficiency, represented by the ratio of neuron covered surface area on the pattern/the total neuron covered surface area, was higher than 90%. Thus, the guiding was significantly enhanced in comparison to earlier reports based on the optimized composition of particles and particle backfill. The guiding efficiency was determined accordingly to a previously reported procedure.56
3. Discussion
In this report we describe the tuning of the chemical composition of solid surfaces on the nanometer scale by controlling the density of AF-AuNP and their surrounding matrix. The amino-functionalized nanoparticles act as anchor point for the attachment of cell adhesion ligands. The successful immobilization and modification of the nanoparticles has been evaluated by SEM and XPS, respectively. Together with the observation that the particles remain in place even after extended culture time proves that backfilling from vapor-phase does not impair the binding of AF-AuNP.Since the density of surface associated particles can be varied, we are able to control the density of binding cues on the surface. This has a significant impact on the viability of the neurons. At the lowest densities only a few particles (surface coverage of 0.1%) can be observed on an area of 1 × 1 μm2. However, each of these particles carries several thousand aminoalkyl thiol molecules. Assuming a coverage of 5.5 AUT per nm2 (ref. 75) on the particle surface we obtain a ligand density of 3.1 × 103 AUT molecules per particle if we take only the upper, solution exposed semi-sphere of the particle into account. The Debye length is about 1 nm for cell culture media, which leads to an effective screening of electrostatic interactions. Therefore, we can assume that the range of interactions of an individual particle is restricted to its hydrodynamic radius of about 11 nm. Consequently, only a small number of highly charged but locally confined adhesion spots are available at sample surfaces with low particle densities. Astonishingly, a relatively high number of vital neurons per unit area was observed for SiO2 samples with low particle densities and without backfill. This indicates that the neurons manage to adhere at particle free areas of the SiO2 surface. This assumption was confirmed by the spreading of neurons from particle containing to particle free areas, Fig. 3. The presence of particle associated adhesion ligands enhances the neuron viability, however is not mandatory for cell survival. The isoelectric point of silicon dioxide can be typically found at pH values of 2–3.76 Thus, it can be assumed that the SiO2 surface carries a negative net charge at cell culture conditions. It has been reported that the adhesion of cells to negatively charged surfaces is rather limited due to electrostatic repulsion with the cell membrane, which is negatively charged too (−18 × 10−3 C m−2).77 However, it is also an established fact that the hydrophilic properties of the SiO2 surface support the adsorption of solution-borne substances,1 which may originate from the neurons themselves. For instance neurotrophins and growth factors can get associated to negatively charged glycoforms of cell adhesion molecules. Such solution-borne species apparently impair the cell aversive properties of SiO2.
Applying a backfill to the particle environment improves the control over the surface composition by preventing undesired adsorption of interfering binding cues. The effectiveness of the 2-[methoxy(polyethyleneoxy)6-9-propyl] trichlorosilane and octyltrichlorosilane backfill coating was demonstrated by particle free control experiments where a clear suppression of neuron adhesion was achieved. The viability of neurons is very low even for samples modified by AF-AuNP at density lower than 30 particles per μm2 in the presents of PEG-based backfills. The number of surface tethered adhesion ligands is not sufficient to support maturation to vital neurons. With increasing particle density, the relative live cell density rises until it levels off at about 1.5 to 1.7. Under these saturation conditions, neurons exhibit a matured development with long as well short, branched neurites at 5 DIV for no backfill and PEG silane backfill. The size of the soma, number of neurite processes, as well as the length of the axons are similar to neurons plated on a PDL control culture and correspond well to a neuronal development of stage 4.73 It can be assumed that the neurons find a sufficient number of surface associated binding cues for their development. Also samples modified with OTS backfill reaches neuron densities values similar to PEG silane and no backfilled samples at high AF-AuNP densities. It is remarkable, that we find about the same relative live cell density for particle densities higher than 500 particles per μm2, independently of the employed backfill. Together with the observation that cell density asymptotically converges towards this value suggest that the number of receptors that get associated to the particles saturates.
To better understand the nature of the electrostatic interactions between cell and sample surface, we estimated the cationic charge density of the sample by multiplying the number of aminoalkyl thiol adsorbed to one AuNP with the respective particle density. We assume that the backfill prevents the adsorption of charged, solution-borne compounds. Considering only the AF-AuNP associated ligands, we obtain a surface charge density of 24 × 10−2 C m−2 for 500 particles per μm2. This value is about one order of magnitude higher than the surface charge density of 15 × 10−3 C m−2 determined for HEK293 cells.3 The difference between sample associated positive charges and negative cell surface charges suggests that an excess of positive charges is required at the sample surface to support the neuron adhesion. In fact, a particle density of about 30 AuNP per μm2 would be sufficient to compensate surface charge density of a cell, however are not sufficient to achieve a saturation of the live cell density, Fig. 2. One possible reason for this finding is the unequal distribution of charges on both surfaces. In the case of AuNP, a large number of amino-ligands (see above) is bound to a single anchor spot with a hydrodynamic radius of 11 nm. The short Debye length of the cell culture media leads to an effective screening of electrostatic interactions. On the contrary, the density of cell adhesion molecules (CAMs) on the cell membrane is in the range of 500 to several thousand per square micrometer.22 The CAMs exhibit glycoforms with attached glycans. These glycans can carry a high number of negative charges as in the case of NCAM associated polysalic acid (PSA). However, also these charged entities are screened by the high concentration of salt ions which confines the range of possible electrostatic interactions. Due to the ionic screening under culture conditions just a limited number of the CAMs would be able to bind to the surface if the particle density is as low as 30 AuNP per μm2. This suggests that a certain number of homogeneously distributed AuNP is required to facilitate a sufficient binding of glycosylated CAMs which finally enables good cell viability. This is seemingly the case at 500 randomly distributed 20 nm AuNP per μm2 for all types tested backfill, which is in the same order with the approximate density of CAMs on the cell membrane. Although the size of the particles is in the same range of the most glycosylated proteins it cannot be excluded that several proteins bind to the same particle. Furthermore, it is likely that several types of glycosylated CAMs are involved in the binding process, since the electrostatic interactions are rather unspecific.
Although we found similar maximum cell viability for all types of particle background we observed distinct differences for the maturation of the cells. In particular the development of neurites is different for samples with OTS backfill in comparison to the other samples. The hydrophobic alkyl-silanes effectively suppress neuronal adhesion and development even at relatively high densities of AF-AuNP (300 particles per μm2). At maximum particle densities (500 particles per μm2) we find neuron viability and number of neurites similar to samples with no or PEG backfill but the length of the axons is significantly reduced to about 1/3. The tendency to develop shorter neurite processes is probably associated to the hydrophobic passivation of the particle surrounding matrix for protein adsorption. Apparently neuron adhesion and neurite outgrowth depend differently onto the chemical background of cationic adhesion ligands. A selective dependence of axonal features on particle density has been recently observed also by Thelen et al. for dorsal root ganglia (DRG).53 During path finding as well as for adhesion the neuron can integrate over large distances and overgrow cell aversive sample areas with the length of several tens of microns.7 However, small neurite extensions which possess often a width smaller than 200 nm, Fig. 5, are more sensitive to variations of the surface composition on nanometer scale.
It turns out that the control of the composition of particle surrounding area is a powerful tool to tune the properties of surfaces on nanometer scale. The influence of the particle composition on the local neuron adhesion and neurite outgrowth was demonstrated previously.56 However, a high guiding accuracy is hampered by an undefined composition of the particle surrounding. On average, we find a live cell density of 30% at bare SiO2 without surface tethered adhesion ligands in comparison to samples with high particle densities. The composition of backfill free SiO2 surface areas may change over time due to adsorption, desorption, and surface diffusion of undefined solution-borne compounds.78 This may strongly affect the reproducibility of the neuron adhesion properties of samples without backfill and has direct impact on guiding attempts. The adhesion contrast between particle free and particle decorated areas is only moderate.
The backfill increases the adhesion contrast for the neuron adhesion. Here, we define the adhesion contrast (C) as ratio of the difference between relative live cell density of cell attractive surface (A) and relative live cell density of cell aversive surface (R) divided by the relative live cell density of cell attractive surface, eqn (1)
| C = (A − R)/A. | (1) |
A value of C = 0 indicates no difference in adhesion between the two surface domains whereas C = 1 represents a perfect adhesion contrast. This cell type specific qualitative parameter allows to estimate the appropriateness of a material boarder to restrict the spreading of cells across this boarder. From Fig. 2, we determined a chemical contrast of C = 0.53 for AF-AuNP on backfill free SiO2; a value of C = 0.87 for AF-AuNP with PEG backfill; and a value of C = 1 for AF-AuNP on OTS modified SiO2. Therefore we used the relative live cell density at 500 particle per μm2 as A and the relative live cell density at 10 particle per μm2 for the respective types of backfill as R. The latter represents the averaged particle density observed for areas between the particle patterns, Fig. 6.66 The high contrast obtained by the application of a backfill resulted in an improvement of the guiding efficiency from 71% for backfill free nanoparticle patterns to 92% for backfilled but otherwise identical surfaces. It should be noted, that the high adhesion contrast was achieved for a surface that was entirely covered with particles. Solely the density of the particles varies from 10 particle per μm2 to 500 particle per μm2. The local density and distribution of nanometer sized domains of cell attractive and cell aversive cues controls the adhesion of cortical neurons and their maturation.
4. Experimental section
4.1 Self-assembling AuNPs
A Si–SiO2 wafer was cut into pieces of 10 × 10 mm2 (〈100〉, n-doped, 1–10 Ω cm, 100 nm-thick dry thermal grown silicon dioxide). The substrates were sequentially cleaned with acetone (Sigma) and 2-propanol (Sigma-Aldrich) in an ultrasonic bath for 3 minutes respectively, followed by rinsing with copious of Milli-Q water, then dried with N2 flow. The cleaned substrates were activated with oxygen plasma at 200 W for 2 min (100-E plasma system, Technics Plasma GmbH). Thereafter, a monolayer of 3-aminopropyltriethoxysilane (APTES, 99%, Sigma) was formed on the surface by using vapor deposition in a glove box. Citrate stabilized AuNPs (0.01 wt%, Sigma) were deposited onto the substrates overnight.4.2 Ligand immobilization
11-Amino-1-undecanethiol hydrochloride (AUT) (Sigma) was dissolved in absolute ethanol. After removal of citrate with oxygen plasma for 2 min, the substrate was soaked into 0.5 mM ligand solution overnight. Afterwards, the substrate was rinsed with copious of Milli-Q water and dried with N2-flow before further use. For the modification of the nanoparticle background, the samples are modified with octyltrichlorosilane or 2-[methoxy(polyethyleneoxy)6-9-propyl] trichlorosilane right after citrate removal. Before ligand modification, the substrate was washed with dehydrated toluene and ethanol to remove any physically absorbed silane molecules. Then, the substrates were modified with 0.5 mM AUT dissolved in ethanol.4.3 Characterization
The composition of the substrate was investigated with XPS to evaluate binding of the 11-amino-1-undecanethiol linked to gold nanoparticles, (PHI 5000 VersaProbe II, ULVAC-PHI). The silicon substrates were washed with acetone, 2-propanol and Milli-Q water, dried with N2. Right after activation with oxygen plasma for 2 min, the samples were silanized with OTS and PEG silane respectively. The contact angles of the above two surfaces together with the bare silicon substrate were measured with a contact angle measurement system (contact angle measurement instrument OCA 20, Dataphysics) by the Sessile drop method with 5 μL Milli-Q water to estimate their wetting properties.4.4 Cell culture
Rat embryonic cortical neurons were obtained and cultured as described by Brewer et al. with slight modification.79 Embryos were dissected from pregnant Wistar rat at 18 days gestation. Cortices were dissected from the embryonic brains and mechanically dissociated by trituration in 1 mL ice-cold HBSS (without calcium and magnesium), 0.035% sodium bicarbonate, 1 mM pyruvate, 10 mM HEPES (Sigma, Germany), 20 mM glucose (Sigma, Germany), pH 7.4, with a fire-polished, silanized Pasteur pipette. The cell suspension was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 in HBSS (with calcium and magnesium), 0.035% sodium bicarbonate, 1 mM pyruvate, 10 mM HEPES (Sigma), 20 mM glucose (Sigma), pH 7.4. Nondispersed tissue was allowed to settle for 3 min and the resulting supernatant was centrifuged at 200 g for 2 min. The pellet was resuspended in 1 mL neurobasal medium with 1% B27, 0.5 mM glutamine, and getamycin per hemisphere. A fraction of the cells were counter stained with trypan blue and counted in a Neubauer counting chamber. The remaining cells were diluted in supplemented neurobasal medium. Diluted cells were plated onto the substrates at 10
2 in HBSS (with calcium and magnesium), 0.035% sodium bicarbonate, 1 mM pyruvate, 10 mM HEPES (Sigma), 20 mM glucose (Sigma), pH 7.4. Nondispersed tissue was allowed to settle for 3 min and the resulting supernatant was centrifuged at 200 g for 2 min. The pellet was resuspended in 1 mL neurobasal medium with 1% B27, 0.5 mM glutamine, and getamycin per hemisphere. A fraction of the cells were counter stained with trypan blue and counted in a Neubauer counting chamber. The remaining cells were diluted in supplemented neurobasal medium. Diluted cells were plated onto the substrates at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2. The medium was exchanged twice every week. All reagents were purchased from Invitrogen unless otherwise noted.
000 cells per cm2. The medium was exchanged twice every week. All reagents were purchased from Invitrogen unless otherwise noted.
4.5 Immunochemistaining
Cells were characterized by fluorescence microscopy after live dead staining and antibody staining. For live dead staining, cells were incubated in a solution of calcein AM (live cells staining) and ethidium homodimer (dead cells staining) in PBS for 10 min. For antibody staining, cells were rinsed twice with pre-warmed PBS (pH 7.4) at 37 °C, and fixed with 4% paraformaldehyde solution for 7 min at 4 °C. After rinsing cells for three times with PBS, the fixed cells were permeabilized with 0.1% Triton X in blocking buffer for 15 min at room temperature and were again washed three times with PBS. Non-specific binding sites were blocked with 2% bovine serum albumin (BSA) and 2% goat serum in PBS overnight. To distinguish neurons from glia cells, the samples were immersed into mouse anti-MAP2 primary antibody (neuron specific) and rabbit anti-Glial Fibrillary Acidic Protein (GFAP) primary antibody with a working concentration of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in BSA blocking solution for 90 min and washed three times with PBS. Subsequently, the cells were incubated with secondary antibody (Alexa Fluor 488 anti-mouse and Alexa Fluor 568 anti-rabbit) together with DAPI for another 90 min. Finally, the substrates were washed three times with PBS and once with water before mounting in Dako Fluorescent mounting medium. Fluorescent imaging of stained samples was carried out with an AxionInager Z1 (Zeiss). Images were taken with an AxioCam MRm camera (Zeiss) and processed using AxioVision 4.7 software (Zeiss). In the case of axon and dendrite specific staining, rabbit anti-MAP2 primary antibody and mouse anti-Tau1 primary antibody were used. Alexa Fluor 488 anti-mouse (Invitrogen) and Alexa Fluor 568 anti-rabbit (Invitrogen) were used as second antibody. The remaining processes were the same as for neurons and glials staining. Neurolucida software was used for cells counting and cell viability measurements.
500 in BSA blocking solution for 90 min and washed three times with PBS. Subsequently, the cells were incubated with secondary antibody (Alexa Fluor 488 anti-mouse and Alexa Fluor 568 anti-rabbit) together with DAPI for another 90 min. Finally, the substrates were washed three times with PBS and once with water before mounting in Dako Fluorescent mounting medium. Fluorescent imaging of stained samples was carried out with an AxionInager Z1 (Zeiss). Images were taken with an AxioCam MRm camera (Zeiss) and processed using AxioVision 4.7 software (Zeiss). In the case of axon and dendrite specific staining, rabbit anti-MAP2 primary antibody and mouse anti-Tau1 primary antibody were used. Alexa Fluor 488 anti-mouse (Invitrogen) and Alexa Fluor 568 anti-rabbit (Invitrogen) were used as second antibody. The remaining processes were the same as for neurons and glials staining. Neurolucida software was used for cells counting and cell viability measurements.
4.6 Focus ion beam cutting and scanning electronic microscopy characterization
Cells were fixed for focus ion beam (FIB) cutting as follows: cells were rinsed twice with pre-warmed PBS (37 °C) and then fixed with 3.2% glutaraldehyde in PBS (pH 7.4) for 2 h. Samples were rinsed twice with PBS and subsequently dehydrated with increasing concentration of ethanol (30%, 50%, 70% for 10 min respectively, 90%, 95%, 100% each 5 min for three times, 100% for storage). The FIB cutting was done with a Helios Nanolab 600i apparatus (FEI). Cells were embedded in a 1 μm-thick platinum layer and subsequently cut by a Ga-ion beam at a beam current of 2 nA and an acceleration voltage of 30 kV. SEM images were performed by Gemini 1550 (Leo/Zeiss) using an inlens detector. SEM images with different particle densities were processed and counted with the aid of ScionImage software.665. Conclusions
In summary, we demonstrate in this work that both neuron viability and maturation can be efficiently influenced not only by changing the density of gold nanoparticles on a SiO2 surface but also alternating the chemical composition of their environment on nanometer scale. The AuNP were modified by aminoalkyl thiol molecules and act as adhesion spots for the immobilization of neurons. The particle surrounding was altered from hydrophilic SiO2, to protein aversive poly(ethylene glycol)-molecules, and hydrophobic alkylsilane molecules. Increasing densities of amino-functionalized nanoparticles enhance the adhesion of vital neurons and saturate at about 500 AuNP per μm2 which is in the same order as the density of glycosylated cell adhesion molecules on cell membranes. The density of the particle associated positive charges is about one order of magnitude larger than the previously reported surface charge density of the membrane glycocalyx. Nevertheless, the viability of neurons on these samples is higher than on homogenously covered PLL samples. Moreover, the backfill also influences the development of neurites on the surface. In particular the length of the neurites was reduced by 2/3 for samples with hydrophobic backfill in comparison to samples with hydrophilic particle environment. By modifying the particle's surrounding matrix, the cell adhesion can be suppressed for low particle densities but allows healthy neuron maturation for dense particle packing. The enhanced chemical contrast between particle free and particle containing areas resulted in an increase of the guiding efficiency for rat cortical neurons to more than 90% in contrast to previous results.By means of surface tethered nanoparticles the composition of solid surfaces can be tuned on nanoscale to match the number and distribution of various types of adhesion molecules in the cell membrane. This approach can contribute to the investigation the complex processes involved in neuron adhesion on the scale of adhesion complexes and to meet demanding challenges of neuroscience and bioelectronics such as inducing polarity, controlling neuronal co-culture, and prevention of glia proliferation. Moreover, this technique can be used as versatile tool to engineer technical surfaces to enhance cell viability, reduce immune responses, and improve the cell/device communication of bioelectronic hybrid systems.
Acknowledgements
We kindly thank E. Brauweiler for FIB investigations and F. Santoro for helpful discussions on FIB cut preparations. We thank C. Kaminski for XPS characterization. M. Prömpers and B. Breuer are appreciated for kindly providing us with the silicon dioxide wafer and cells, respectively. P. Li is grateful to Chinese scholarship council for predoctoral stipends.Notes and references
- P. Roach, T. Parker, N. Gadegaard and M. R. Alexander, Surf. Sci. Rep., 2010, 65, 145 CrossRef CAS.
- J. M. Peyrin, B. Deleglise, L. Saias, M. Vignes, P. Gougis, S. Magnifico, S. Betuing, M. Pietri, J. Caboche, P. Vanhoutte, J. L. Viovy and B. Brugg, Lab Chip, 2011, 11, 3663 RSC.
- K. Riehemann, S. W. Schneider, T. A. Luger, B. Godin, M. Ferrari and H. Fuchs, Angew. Chem., Int. Ed., 2009, 48, 872 CrossRef CAS PubMed.
- G. J. Chader, J. Weiland and M. S. Humayun, in Neurotherapy: Progress in Restorative Neuroscience and Neurology, Progress in Brain Research, ed. J. Verhaagen, E. M. Hol, I. Huitenga, J. Wijnholds, A. B. Bergen, G. J. Boer and D. F. Swaab, Elsevier Science BV, Amsterdam, Netherlands, 2009, vol. 175, pp. 317–332 Search PubMed.
- W. M. Grill, S. E. Norman and R. V. Bellamkonda, Annu. Rev. Biomed. Eng., 2009, 1 CrossRef CAS PubMed.
- A. Poghossian, S. Ingebrandt, A. Offenhäusser and M. J. Schöning, Semin. Cell Dev. Biol., 2009, 20, 41 CrossRef CAS PubMed.
- A. Offenhäusser, S. Böcker-Meffert, T. Decker, R. Helpenstein, P. Gasteier, J. Groll, M. Möller, A. Reska, S. Schäfer, P. Schulte and A. Vogt-Eisele, Soft Matter, 2007, 3, 290 RSC.
- P. K. Mattila and P. Lappalainen, Nat. Rev. Mol. Cell Biol., 2008, 9, 446 CrossRef CAS PubMed.
- C. Conde and A. Caceres, Nat. Rev. Neurosci., 2009, 10, 319 CrossRef CAS PubMed.
- D. A. Fletcher and D. Mullins, Nature, 2010, 463, 485 CrossRef CAS PubMed.
- F. Polleux and W. Snider, Cold Spring Harbor Perspect. Biol., 2010, 2, a001925 CAS.
- D. Kleinfeld, K. H. Kahler and P. E. Hockberger, J. Neurosci., 1988, 8, 4098 CAS.
- B. F. Liu, J. Ma, E. J. Gao, Y. He, F. H. Cui and Q. Y. Xu, Biosens. Bioelectron., 2008, 23, 1221 CrossRef CAS PubMed.
- Y. Naka, A. Eda, H. Takei and N. Shimizu, J. Biosci. Bioeng., 2002, 94, 434 CrossRef CAS PubMed.
- A. P. Balgude, X. Yu, A. Szymanski and R. V. Bellamkonda, Biomaterials, 2001, 22, 1077 CrossRef CAS PubMed.
- L. A. Flanagan, Y. E. Ju, B. Marg, M. Osterfield and P. A. Janmey, NeuroReport, 2002, 13, 2411 CrossRef PubMed.
- H. G. Sundararaghavan, G. A. Monteiro, B. L. Firestein and D. I. Shreiber, Biotechnol. Bioeng., 2009, 102, 632 CrossRef CAS PubMed.
- G. A. Dunn and J. P. Heath, Exp. Cell Res., 1976, 101, 1 CrossRef CAS PubMed.
- J. S. Goldner, J. M. Bruder, G. Li, D. Gazzola and D. Hoffman-Kim, Biomaterials, 2006, 7, 460 CrossRef PubMed.
- A. M. Rajnicek, S. Britland and C. D. McCaig, J. Cell Sci., 1997, 110, 2905 Search PubMed.
- T. Wolfram, F. Belz, T. Schoen and J. P. Spatz, Biointerphases, 2007, 2, 44 CrossRef CAS PubMed.
- T. Wolfram, J. P. Spatz and R. W. Burgess, BMC Cell Biol., 2008, 9, 64 CrossRef PubMed.
- J. Mai, et al. , J. Neurosci., 2009, 29, 7450 CrossRef CAS PubMed.
- S. W. Moore, X. Zhang, C. D. Lynch and M. P. Sheetz, J. Neurosci., 2012, 32, 11574 CrossRef CAS PubMed.
- A. C. von Philipsborn, S. Lang, J. Loeschinger, A. Bernard, C. David, D. Lehnert, F. Bonhoeffer and M. Bastmeyer, Development, 2006, 133, 2487 CrossRef CAS PubMed.
- M. Arnold, M. Schwieder, J. Blümmel, E. A. Cavalcanti-Adam, M. López-García, H. Kessler, B. Geiger and J. P. Spatz, Soft Matter, 2009, 5, 72 RSC.
- R. G. Ellis-Behnke, Y. X. Liang, S. W. You, D. K. C. Tay, S. G. Zhang, K. F. So and G. E. Schneider, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 5054 CrossRef CAS PubMed.
- C. Staii, C. Viesselmann, J. Ballweg, L. Shi, G. Liu, J. C. Williams, E. W. Dent, S. N. Coppersmith and M. A. Eriksson, Biomaterials, 2009, 30, 3397 CrossRef CAS PubMed.
- A. K. Vogt, L. Lauer, W. Knoll and A. Offenhäusser, Biotechnol. Prog., 2003, 19, 1562 CrossRef CAS PubMed.
- A. K. Vogt, G. J. Brewer, T. Decker, S. Böcker-Meffert, V. Jacobsen, M. Kreiter, W. Knoll and A. Offenhäusser, Neuroscience, 2005, 134, 783 CrossRef CAS PubMed.
- P. B. van Wachem, T. Beugeling, J. Feijen, A. Bantjes, J. P. Detmers and W. G. van Aken, Biomaterials, 1985, 6, 403 CrossRef CAS PubMed.
- J. H. Lee, H. W. Jung, I. K. Kang and H. B. Lee, Biomaterials, 1994, 15, 705 CrossRef CAS PubMed.
- E. Yavin and Z. Yavin, J. Cell Biol., 1974, 62, 540 CrossRef CAS PubMed.
- M. Matsuzawa, R. S. Potember and V. Krauthamer, Brain Res., 1994, 667, 47 CrossRef CAS PubMed.
- M. Matsuzawa, R. S. Potember, D. A. Stenger and V. Krauthamer, J. Neurosci. Methods, 1993, 50, 253 CrossRef CAS PubMed.
- P. M. S. John, L. Kam, S. W. Turner, H. G. Craighead, M. Issacson, J. N. Turner and W. Shain, J. Neurosci. Methods, 1997, 75, 171 CrossRef.
- S.-J. Yen, W.-L. Hsu, Y.-C. Chen, H.-C. Su, Y.-C. Chang, H. Chen, S.-R. Yeh and T.-R. Yew, Biosens. Bioelectron., 2011, 26, 4124 CrossRef CAS PubMed.
- R. F. Valentini, A. M. Sabatini, P. Dario and P. Aebischer, Brain Res., 1989, 480, 300 CrossRef CAS PubMed.
- C. P. Johnson, I. Fujimoto, U. Rutishauser and D. E. Leckband, J. Biol. Chem., 2005, 280, 137 CrossRef CAS PubMed.
- A. Blau, Curr. Opin. Colloid Interface Sci., 2013, 18, 481 CrossRef CAS.
- M. Arnold, E. A. Cavalcanti-Adam, R. Glass, J. Blummel, W. Eck, M. Kantlehner, H. Kessler and J. P. Spatz, ChemPhysChem, 2004, 5, 383 CrossRef CAS PubMed.
- D. Du, S. L. Liu, J. Chen, H. X. Ju, H. Z. Lian and J. X. Li, Biomaterials, 2005, 26, 6487 CrossRef CAS PubMed.
- M. M. Stevens and J. H. George, Science, 2005, 310, 1135 CrossRef CAS PubMed.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk and R. O. Oreffo, Nat. Mater., 2007, 6, 997 CrossRef CAS PubMed.
- M. Kim, M. Park, K. Kanga and I. S. Choi, Biomater. Sci., 2014, 2, 148 RSC.
- S. Xie and R. Luttge, Microelectron. Eng., 2014, 124, 30 CrossRef CAS.
- A. P. Alivisatos, A. M. Andrews, E. S. Boyden, M. Chun, G. M. Church and K. Deisseroth, et al. , ACS Nano, 2013, 7, 1850 CrossRef CAS PubMed.
- D. Schwaab, P. Zentis, S. Winter, S. Meffert, A. Offenhäusser and D. Mayer, Jpn. J. Appl. Phys., 2013, 52, 05DA19 CrossRef.
- J. H. Huang, S. V. Grater, F. Corbellinl, S. Rinck, E. Bock, R. Kemkemer, H. Kessler, J. D. Ding and J. P. Spatz, Nano Lett., 2009, 9, 1111 CrossRef CAS PubMed.
- M. Arnold, V. C. Hirschfeld-Warneken, T. Lohmuller, P. Heil, J. Blummel, E. A. Cavalcanti-Adam, M. Lopez-Garcia, P. Walther, H. Kessler, B. Geiger and J. P. Spatz, Nano Lett., 2008, 8, 2063 CrossRef CAS PubMed.
- E. A. Cavalcanti-Adam, T. Volberg, A. Micoulet, H. Kessler, B. Geiger and J. P. Spatz, Biophys. J., 2007, 92, 2964 CrossRef CAS PubMed.
- C. Selhuber-Unkel, T. Erdmann, M. Lopez-Garcia, H. Kessler, U. S. Schwarz and J. P. Spatz, Biophys. J., 2010, 98, 543 CrossRef CAS PubMed.
- K. Thelen, S. Jaehrling, J. P. Spatz and G. E. Pollerberg, PLoS One, 2012, 7, e40493 CAS.
- N. Nath and A. Chilkoti, Anal. Chem., 2004, 76, 5370 CrossRef CAS PubMed.
- Y. Guan, M. Li, K. Dong, J. Ren and X. Qu, Small, 2014, 10, 3655 CrossRef CAS PubMed.
- S. Gilles, S. Winter, K. E. Michael, S. H. Meffert, P. G. Li, K. Greben, U. Simon, A. Offenhausser and D. Mayer, Small, 2012, 8, 3357 CrossRef CAS PubMed.
- N. Schwierz, D. Horinek, S. Liese, T. Pirzer, B. N. Balzer, T. Hugel and R. R. Netz, J. Am. Chem. Soc., 2012, 134, 19628 CrossRef CAS PubMed.
- J. Blummel, N. Perschmann, D. Aydin, J. Drinjakovic, T. Surrey, M. Lopez-Garcia, H. Kessler and J. P. Spatz, Biomaterials, 2007, 28, 4739 CrossRef PubMed.
- T. G. Ruardy, H. E. Moorlag, J. M. Schakenraad, H. C. VanderMei and H. J. Busscher, J. Colloid Interface Sci., 1997, 188, 209 CrossRef CAS.
- S. N. M. Dowell-Mesfin, M.-A. Abdul-Karim, A. M. P. Turner, S. Schanz, H. G. Craighead, B. Roysam, J. N. Turner and W. Shain, J. Neural Eng., 2004, 1, 78 CrossRef PubMed.
- A. Vogt, F. D. Stefan, A. Best, G. Nelles, A. Yasuda, W. Knoll and A. Offenhäusser, J. Neurosci. Methods, 2004, 134, 191 CrossRef PubMed.
- S. Zhong, W. E. Teo, X. Zhu, R. W. Beuerman, S. Ramakrishna and L. Y. L. Yung, J. Biomed. Mater. Res., Part A, 2006, 79, 456 CrossRef PubMed.
- K. Czöndör, M. Garcia, A. Argento, A. Constals, C. Breillat, B. Tessier and O. Thoumine, Nat. Commun., 2013, 4, 1 Search PubMed.
- M. S. Ravenscroft, K. E. Bateman, K. M. Shaffer, H. M. Schessler, D. R. Jung and T. W. Schneider, et al. , J. Am. Chem. Soc., 1998, 120, 12169 CrossRef CAS.
- R. S. Kane, S. Tamayama, E. Ostuni, D. E. Ingber and G. M. Whitesides, Biomaterials, 1999, 20, 2363 CrossRef CAS PubMed.
- S. Gilles, C. Kaulen, M. Pabst, U. Simon, A. Offenhausser and D. Mayer, Nanotechnology, 2011, 22, 295301 CrossRef CAS PubMed.
- M. A. Dyer, K. M. Ainslie and M. V. Pishko, Langmuir, 2007, 23, 7018 CrossRef CAS PubMed.
- P. V. Ganesan and S. G. Boxer, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 5627 CrossRef CAS PubMed.
- S. R. Wasserman, Y. T. Tao and G. M. Whitesides, Langmuir, 1989, 5, 1074 CrossRef CAS.
- S. Saneinejad and M. S. Shoichet, J. Biomed. Mater. Res., 1998, 42, 19 CrossRef.
- Y. Nam, J. C. Chang, B. C. Wheeler and G. J. Brewer, IEEE Trans. Biomed. Eng., 2004, 51, 165 CrossRef PubMed.
- N. E. Sanjana and S. B. Fuller, J. Neurosci. Methods, 2004, 136, 163 CrossRef PubMed.
- S. Kaech and G. Banker, Nat. Protoc., 2006, 1, 2406 CrossRef CAS PubMed.
- H. Lee, Y. Jang, J. Seo, J. M. Nam and K. Char, ACS Nano, 2011, 5, 5444 CrossRef CAS PubMed.
- H. Hinterwirth, S. Kappel, T. Waitz, T. Prohaska, W. Lindner and M. Lammerhofer, ACS Nano, 2013, 7, 1129 CrossRef CAS PubMed.
- M. Kosmulski, J. Colloid Interface Sci., 2011, 353, 1 CrossRef CAS PubMed.
- P. C. Zhang, A. M. Keleshian and F. Sachs, Nature, 2001, 413, 428 CrossRef CAS PubMed.
- T. Blaudeck, P. A. Ersman, M. Sandberg, S. Heinz, A. Laiho, J. Liu, I. Engquist, M. Berggren and R. R. Baumann, Adv. Funct. Mater., 2012, 22, 2939 CrossRef CAS.
- G. J. Brewer, J. R. Torricelli, E. K. Evege and P. J. Price, J. Neurosci. Res., 1993, 35, 567 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |