Comparison of hydrocarbon-and lactam-bridged cyclic peptides as dimerization inhibitors of Leishmania infantum trypanothione reductase†
Pedro A. Sánchez-Murciaab,
Marta Ruiz-Santaquiteriab,
Miguel A. Toroc,
Héctor de Lucioc,
María Ángeles Jiménezd,
Federico Gagoa,
Antonio Jiménez-Ruizc,
María-José Camarasab and
Sonsoles Velázquez*b
aDepartamento de Ciencias Biomédicas, Unidad Asociada al CSIC, Universidad de Alcalá, Madrid, Spain
bInstituto de Química Médica (IQM-CSIC), Madrid, Spain. E-mail: iqmsv29@iqm.csic.es; Fax: +34 91 5644853; Tel: +34 91 2587458
cDepartamento de Biología de Sistemas, Universidad de Alcalá, Madrid, Spain
dInstituto Química-Física Rocasolano (IQFR-CSIC), Madrid, Spain
First published on 19th June 2015
Abstract
All-hydrocarbon and lactam-bridged staples linking amino acid side-chains have been used to stabilize the α-helical motif in short 13-mer peptides that target critical protein–protein interactions at the dimerization interface of Leishmania infantum trypanothione reductase (Li-TryR). The design of the best positions for covalent hydrocarbon closure relied on a theoretical prediction of the degree of helicity of the corresponding cyclic peptides in water. Selected (i, i + 4) and (i, i + 7) hydrocarbon-stapled peptides were prepared by using solid-phase synthesis protocols and optimized ring-closing metathesis reactions under microwave conditions. Structural analysis by NMR spectroscopy confirmed high helical contents in aqueous TFE solutions for both types of helix-constrained cyclic peptides. Remarkably, the ability to prevent Li-TryR dimerization was reduced in both (i, i + 4) and (i, i + 7) hydrocarbon stapled peptides but was retained in the corresponding (i, i + 4) Glu–Lys lactam-bridged analogue, which also showed a higher resistance to proteolytic degradation by proteinase K relative to the linear peptide prototype. In silico studies indicated that the introduction of a hydrocarbon staple vs. a lactam bridge likely perturbs critical interactions required for proper binding of the peptide to the Li-TryR monomer.
Introduction
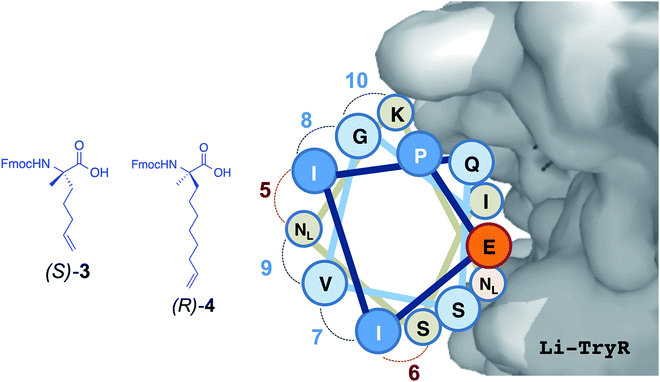
Leishmaniasis is a parasitic infection caused by unicellular protozoan organisms (Leishmania spp.) belonging to the Trypanosomatidae family. Although this disease is seen nowadays as a global health problem,1 currently available drugs are decades old and have several drawbacks including poor efficiency and bioavailability, toxicity to humans, high cost, and the emergence of drug-resistant parasitic strains.2–5 Thus, there is an urgent need to find new targets and/or inhibition mechanisms against these parasites. Trypanothione reductase (TryR; EC 1.8.1.12) is a NADPH flavoenzyme unique to and absolutely essential for protozoan parasites from the genera Trypanosoma and Leishmania. TryR catalyzes the reduction of trypanothione disulfide to the dithiol form.6–8 The cellular role of trypanothione and TryR is preserving a balanced intracellular redox state, a task equivalent to that of the glutathione/glutathione reductase (GR) pair in mammals.6 Because of the lack of catalase and glutathione peroxidase activities, these parasites rely solely on the trypanothione system as a defense mechanism against the oxidative stress that is generated either by their own metabolism or by the host immune response.In addition to playing a pivotal role in the parasite, another characteristic that makes TryR a potential target for antiparasitic drugs is the different substrate specificity from that of GR, the enzyme with the most similar function in humans. These facts make TryR a very attractive validated target for drug development against trypanosomatid-caused infections. Several inhibitors of TryR have been reported including those based on tricyclic antidepressants,9,10 polyamine conjugates11,12 and substrate analogues.9–14 Most of the efforts to inhibit TryR have relied on the design of molecules directed towards the active site but with limited success. Taking advantage of the fact that the functional form of TryR is a homodimer, we have recently reported an alternative inhibition strategy aimed at disrupting the dimer interface of the enzyme by means of protein–protein interaction inhibitors15 (Fig. 1). By a combination of molecular modelling and site-directed mutagenesis studies we identified and validated E436 as a key amino acid (hot spot) for the structural stability and function of the dimer. On the basis of these results and as a “proof of concept” of this novel approach we designed and tested a small library of linear peptides whose sequences were directly derived from the helical homodimerization domain containing E436 (residues P435-M447) of Leishmania infantum TryR (Li-TryR). Among the synthesized peptides, the linear PEIIQSVGISMKM (1) and PEIIQSVGIS-Nle-K-Nle (2) 13-mers, in which a Cys and/or the two Met residues of the wild-type peptide sequence were replaced by isosteric Ser and Nle (in order to prevent potential oxidation), stood out as promising Li-TryR dimerization inhibitors in the micromolar range (Fig. 1).15
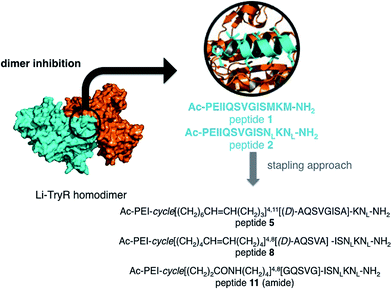 | ||
| Fig. 1 Li-TryR dimer inhibition strategy and designed stapled peptides synthesized in this work. NL: norleucine. | ||
Covalent side-chain to side-chain linkage (“stapling”) within small α-helical segments derived from a protein has allowed to overcome the little or no helical character shown in solution by these oligopeptides when they are excised from their native context.16 Thus, incorporation of both lactam bridges17–19 and short hydrocarbon chains20,21 as staples has been shown to enhance helicity, resistance to proteolysis and cell permeability. To date, numerous examples of stapled peptides with affinity for several therapeutic targets and the ability to modulate protein–protein interactions have been reported.22,23
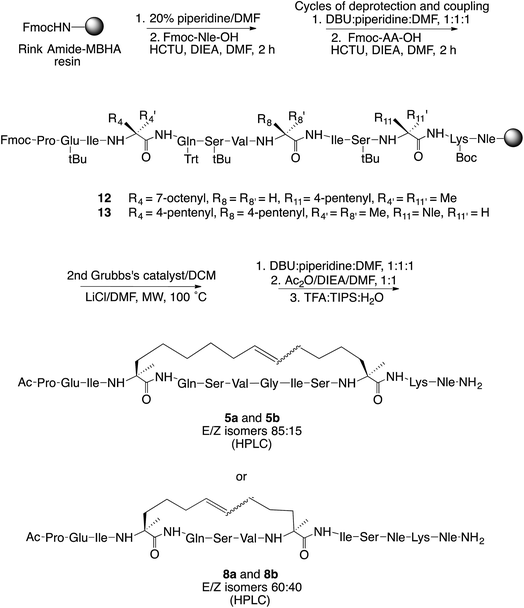
In order to stabilize the α-helical structure of the short linear prototype 2, we herein describe our initial studies on the effects of side-chain hydrocarbon stapling (peptides 5 and 8) [at either one (i, i + 4) or two turns (i, i + 7) of the α-helix] or incorporation of a lactam bridge (peptide 11) (Fig. 1) on (i) the peptide helical conformation, (ii) the proteolytic stability, and (iii) the ability to disrupt Li-TryR dimerization. Our data show that the all-hydrocarbon stapled peptides are inactive in Li-TryR dimerization assays whereas the lactam-bridged cyclic peptide stands out as a potent inhibitor of TryR dimerization. A rationale for these significant differences was provided by analysing the results obtained from molecular dynamics (MD) simulations in explicit solvent.
Results and discussion
In silico design of stapled peptides
From available helix stabilization strategies,16,17 we first selected the all-hydrocarbon stapling approach developed by Verdine and colleagues.21 It is well established that for i, i + 4 or i, i + 7 stapling, the most stereochemically favourable cross-link to stabilise helices is through two units of (S)-α-methyl-α-pentenylglycine (3) to form Si, i+4 S(8)24 staples or between one unit of (R)-α-methyl-α-octenylglycine (4) (figure in Table 1) and one unit of 3 to form Ri,i + 7S(11)25 staples according to the nomenclature described by Verdine et al.21a Since the staple position critically influences the overall degree of structure and biological activity of a given peptide,21a,26 we first performed an in silico analysis to select the best positions for covalent i, i + 4 and i, i + 7 closures that would lead to a higher degree of stabilization of the helical structure in linear peptide 2 (Table 1). All the staples in peptide 2 were placed on the face opposite to that used for binding to the Li-TryR monomer so as to preserve the crucial interactions at the interface.| Peptides | Position | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| a NL: norleucine. | |||||||||||||
| 1 (linear) | P | E | I | I | Q | S | V | G | I | S | M | K | M |
| 2 (prototype) | P | E | I | I | Q | S | V | G | I | S | NL | K | NL |
| 5 | P | E | I | 4 | Q | S | V | G | I | S | 3 | K | NL |
| 6 | P | E | 4 | I | Q | S | V | G | I | 3 | NL | K | NL |
| 7 | P | E | 3 | I | Q | S | 3 | G | I | S | NL | K | NL |
| 8 | P | E | I | 3 | Q | S | V | 3 | I | S | NL | K | NL |
| 9 | P | E | I | I | Q | S | 3 | G | I | S | 3 | K | NL |
| 10 | P | E | I | I | Q | S | V | 3 | I | S | NL | 3 | NL |
Table 1 highlights all possible positions in the sequence of 2 for the introduction of the non-natural α,α-disubstituted residues bearing the double bonds (3 and 4) which would lead to the hydrocarbon bridge after ruthenium-catalyzed ring closing metathesis (stapled peptides 5–10). There are six possible stapled peptides: 5 and 6 (i, i + 7 staples) and 7–10 (i, i + 4 staples).
Stapled peptides 5–10 showed increased degrees of predicted helicity per residue compared to the linear peptide 2 (Fig. 2). In addition, significant differences were found depending on the position and the length of the hydrocarbon staples.
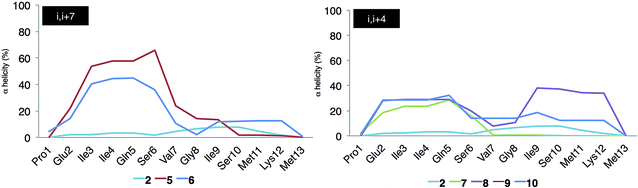 | ||
| Fig. 2 Predicted α-helicities per residue (%) of peptides 5–10 and 2 (prototype) calculated with the dssp algorithm. | ||
In general, the i, i + 7 stapling system was found to be more stabilizing than i, i + 4 in terms of absolute helicity values. Among the possible i, i + 7 staples, peptide 5 displayed the highest α-helix propensity towards the N-terminus, reaching absolute values close to 50% helicity (Fig. 2, in red). On the other hand, peptide 8 with the i, i + 4 staple located at the centre of the helix (Fig. 2, in purple) exhibited the highest α-helix stabilization towards the C-terminus. On the basis of these findings, stapled peptides 5 and 8 (linking positions 4 and 11 and 4 and 8, respectively) were selected for synthesis and further study.
For comparative purposes, the constrained i, i + 4 cyclic analogue with a lactam bridge between the side-chains of Glu–Lys residues at positions 4 and 8 [Ac-PEI(EQSVK)IS-Nle-K-Nle-NH2 (11)] was also selected for study. This enabled us to compare the effects of hydrocarbon versus lactam linkers on helix stabilization and on Li-TryR dimerization disruption.
Chemical synthesis
For hydrocarbon-stapled peptides, we first synthesized the required optically pure α,α-disubstituted amino acids (S)-3 and (R)-4. This was made from the commercially available chiral diphenyloxazinones following the procedure described by Williams and colleagues27 and the modifications reported by Verdine's group.21a In brief, two consecutive asymmetric alkylations on C-3, scaffold cleavage, and final Fmoc protection of the N-terminal amino acid gave the desired quaternary amino acids (S)-3 and (R)-4 (Scheme S1, ESI†).Target stapled peptides 5 and 8 were synthesized manually following the standard Fmoc/tBu solid-phase orthogonal protection strategy on Rink amide-MBHA polystyrene resin (Scheme 1). The appropriate α-methyl-α-alkenylamino acids were introduced at the right positions of the sequence in the elongation step. Dicarba bridge formation was accomplished from the linear precursors 12 and 13 through on-resin ring-closing metathesis (RCM) using Grubb's second-generation ruthenium catalyst. The optimal microwave conditions for the RCM reaction were 100 °C using 20 mol% of catalyst, LiCl as a chaotropic agent28 and CH2Cl2 as the solvent. Next, the N-terminus of the peptides was acetylated and cleaved from the resin under standard conditions.
The crude peptides were purified on reverse phase solid-phase extraction (SPE) cartridges or semipreparative high performance liquid chromatography (HPLC) to give the desired stapled peptides 5 and 8 in 30% and 6% overall yields, respectively. Both E and Z isomers were detected by HPLC in 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 and 60
15 and 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 ratios for 5 and 8, respectively, and could be partially separated. The major isomers 5a and 8a very probably corresponds to the E isomers since the NMR coupling constant 3JHH between the vinyl protons is approximately 13–15 Hz.
40 ratios for 5 and 8, respectively, and could be partially separated. The major isomers 5a and 8a very probably corresponds to the E isomers since the NMR coupling constant 3JHH between the vinyl protons is approximately 13–15 Hz.
Inhibition of Li-TryR dimerization
The effects on dimer stability of the (i, i + 4 or i, i + 7)-hydrocarbon stapled peptides 5a (major isomer), 8a (major isomer), 8a, b (mixture of isomers) and the (i, i + 4) lactam-bridged analogue 11, and the linear prototype peptide 2 were evaluated by using a highly sensitive and versatile in-house ELISA15 (Table 2). These studies revealed that all-hydrocarbon stapled analogues 5a, 8a and 8a, b did not significantly promote disruption of the Li-TryR dimer. In contrast, the lactam-bridged analogue 11 reduced the detection of labeled Li-TryR dimer to the same extent that did their linear counterpart 2. Thus, even when the same positions are stapled (8a or 8a, b vs. 11), the nature of the covalent linkage has a deep impact on the potency of the peptides as Li-TryR dimerization disruptors.| Peptide | IC50a (μM) |
|---|---|
| a Results are representative of three independent experiments each performed in triplicate. >75 indicates that IC50 is higher than 75 μM (maximum assayed). | |
| 2 (linear prototype) | 15.6 ± 1.4 |
| 5a (major isomer; 4,11-hydrocarbon-stapled) | >75 |
| 8a (major isomer; 4,8-hydrocarbon-stapled) | >75 |
| 8a, b (mixture of isomers; 4,8-hydrocarbon-stapled) | >75 |
| 11 (4,8-lactam-bridged) | 24.8 ± 0.4 |
| Mepacrine (control) | >75 |
NMR structural studies
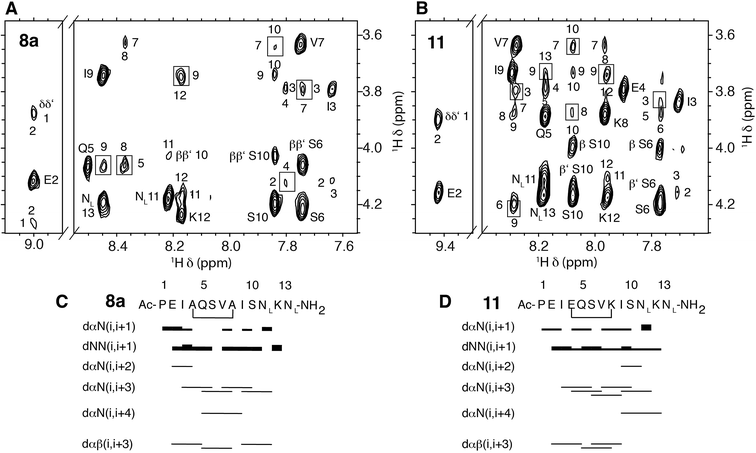
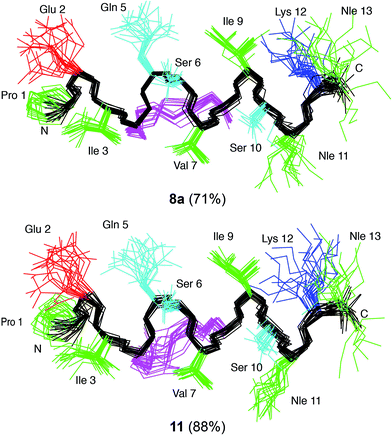
In order to elucidate whether the nature of the covalent linkage could have a differential effect on the structure of the all-hydrocarbon-stapled 8 and lactam-bridged 11 peptides we examined the structural behaviour of these peptides in solution by 1D and 2D NMR spectroscopy. In particular, we selected the lactam-bridged derivative 11 and the major geometric isomer of peptide 8a (isomer E according to a NMR coupling constant 3JHH between the vinyl protons >15 Hz). Both peptides 8a and 11, in the TFE–H2O solvent,29 showed medium-range NOE cross-peaks characteristic of α-helices, i.e. dαN(i, i+2), dαN(i, i+3), dαN(i, i+4), and dαβ(i, i+3) (Fig. 3). As an example, a detail of the 1H,1H-NOESY spectra of 8a and 11 is shown in Fig. 3A and B. Additionally, the deviations of chemical shifts from random coil values shown by the Hα protons (ΔδHα = δHα observed − δHα random coil, ppm) are positive and large in magnitude (Fig. S1†), which are also characteristic of helices. The percentages of helix formed by peptides 8a and 11 in 30% TFE at pH 5.5 and 25 °C are 71% and 88%, respectively, as estimated from the averaged ΔδHα values.30Given the large helical populations presented by stapled peptides 8a and 11, and to visualize their 3D structure, we next performed structure calculations from geometric constraints (distances and dihedral angles; Table S1†) derived from NMR parameters and using the CYANA program.31 The ensemble of the 20 lowest target function conformers resulting for each peptide was well defined, as seen in Fig. 4, and indicated by the pairwise root mean square deviation (RMSD), which is 0.50 ± 0.20 and 0.60 ± 0.20 Å for the backbone atoms of peptides 8a and 11, respectively (Table S1†).
On the other hand, the superimposition of the two best calculated structures for cyclic peptides 8a and 11 with the Li-TryR α-helix Pro435-Met447 in the native protein shows a correct orientation of the side chains of those residues in the interaction face with the protein (Fig. S2†). This confirms our premise that the staple is oriented on the opposite face. However, it is noteworthy that there are no relevant differences in structuring both cyclic peptides as α-helices in TFE/H2O solution, and therefore, the observed variations in the activity of the peptides as dimer disruptors cannot be attributed to differences in the ability of the staples to stabilize the helical structure in solution.
Molecular modelling
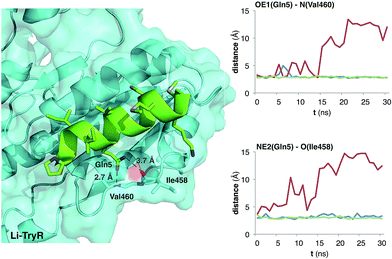
The MD simulation studies (Fig. 5) complemented the NMR solution experiments, as it is known that the conformation of a protein-bound ligand can be different from that found in solution. In this case, we considered both geometric isomers E/Z of all-hydrocarbon stapled peptides 5a, b and 8a, b, the lactam-bridged derivative 11 and the linear prototype 2 (used as a control).We have previously shown15 that the replacement of the Gln side chain at position 5, in similar 13-mer peptides, by Ala has the greatest deleterious effect on their potency as Li-TryR inhibitors. Thus, we monitored the interaction of the former residue with the Li-TryR monomer along the different MD trajectories for all the simulated peptides. As an example, Fig. 5 shows the evolution of the distances between the crucial Gln5 side chain of the peptides and the amide N(Val460) and O(Ile458) backbone atoms of Li-TryR monomer for the hydrocarbon-stapled derivative 8a, the lactam-bridged 11 and the linear counterpart 2. As can be observed, the introduction of the all-hydrocarbon staple in 8a (Fig. 5, in red) perturbs the interaction of Gln5 with the Li-TryR monomer when compared with the linear peptide 2. This is a common feature for the rest of the hydrocarbon-stapled peptides studied along the different MD simulations (data not shown). On the contrary, peptide 11 carrying an amide as the covalent bridge, keeps these interactions to a similar extent as the linear counterpart.
Calculation of the total interaction energies for these peptides, when bound to the Li-TryR monomer along the MD trajectories (Fig. S3 and Table S2†), reveals that the introduction of the all-hydrocarbon staple in both double-bond isomers 5 and 8 results in a significant 30% decrease in the total binding energy relative to the linear peptide 2. Regarding the amide-bridged derivative 11, reduction in the total binding energy is much smaller (14%). These results are in consonance with the experimental IC50 values shown in Table 2. Visual inspection of the representative structures of the major clusters of peptides 2, 8a, b and 11 calculated along the MD trajectories (Fig. S4†) reveals that the unsaturated all-hydrocarbon stapled 8a, b, when bound to the protein, do not preserve the helicity at both N- and C-terminus, in agreement with the fraying observed in the NMR structure of 8a (Fig. 4). Instead, in 11, the amide bridge is compatible with a good orientation of Gln5. Thus, the binding differences observed along the MD trajectories can be attributed to the hydrophobic nature of the hydrocarbon linker. Hydrocarbon stapling technology is generally considered as an effective tool to increase the affinity of α-helical peptides for their targets. However, there are reported examples demonstrating that disruption of stabilizing intramolecular interactions between side chains32 or reduction of the flexibility in the stapled peptide33 can prevent adoption of the conformation needed for efficient binding to their target relative to the unstapled peptide. Thus, our studies complement these previous observations that peptide stapling with a view to increasing helicity does not necessarily result in enhanced binding affinity for the target protein.
Proteolytic stability
Cleavage by proteases is one of the main pathways for inactivation of peptides in a biological setting. Because proteases bind to their substrates in an extended rather than in a helical conformation, increasing or maintaining a helical conformation is expected to confer proteolytic stability. To determine the influence of lactam-bridged stabilization on proteolytic stability of the peptides, we compared the susceptibilities of the linear prototype peptide 2 and the active lactam-bridged analogue 11 towards degradation by proteinase K, a promiscuous serine protease. The half-live for the lactam-bridged analogue 11 was significantly increased, with calculated t1/2 values of 316 min compared to 23 min for the linear peptide 2.34 This 14-fold improvement could be due to the increased α-helical character of the cyclic analogue and/or to the presence of the bridge itself, which would be expected to sterically affect the access to protease-sensitive peptide bonds. This result is consistent with previous findings for helix-constrained peptides35 and makes peptide 11 a new hit that retains the inhibitory potency shown by the linear prototype but it is endowed with improved pharmacokinetic properties. As an illustrative example of all-hydrocarbon stapling, similar proteolytic stability studies with hydrocarbon-stapled analogue 5a revealed comparable improvement in stability compared to the lactam-bridged analogue 11 (t1/2 values of 286 min vs. 316 min, respectively).Conclusions
We have compared two different strategies for covalently linking amino acid side-chains in a short linear peptide (13-mer) that targets a hotspot at the dimerization interface of Li-TryR. A series of rationally designed (i, i + 4) and (i, i + 7) hydrocarbon stapled derivatives linking amino acids at positions 4 and 8 (peptide 5) or 4 and 10 (peptide 8), respectively, and a lactam-bridged analogue of 8 (peptide 11) were investigated. Although the all-hydrocarbon stapling approach has been reported as successful in many published examples, in our case the hydrocarbon-stapled analogues were not able to disrupt the dimerization process of the enzyme whereas the lactam-bridged analogue 11 maintained the potency of the native linear peptide 2. Thus, our biological results reveal that the nature of the covalent linker plays a major role in the peptide's ability to prevent Li-TryR dimerization. Structural NMR studies in solution showed that both hydrocarbon and lactam-bridged peptides 8 and 11 display a high α-helical content in TFE–water mixtures. On the other hand, MD simulations of the linear and cyclic analogues bound on the surface of a Li-TryR monomer indicated that the lactam-bridged peptide 11 could maintain the right orientation of the essential Gln5 side-chain better than the hydrocarbon stapled analogue 8. Thus, our studies represent an example of how the nature of the covalent linkage (hydrocarbon vs. lactam) may perturb critical interactions required for proper binding to the protein. These findings may be taken into account when designing helix-constrained peptides. Interestingly, the lactam-stabilized analogue 11 not only affords the spatial arrangement of the critical residues in the correct conformation for activity but also provides higher proteolytic stability (14-fold) in comparison to its linear peptide counterpart.Experimental
Chemistry
Unless otherwise noted, analytical grade solvents and commercially available reagents were used without further purification. DMF and CH2Cl2 were dried using activated molecular sieves. Water sensitive reactions were carried out under Ar atmosphere. Chiral diphenyloxazinones, Grubb's second-generation ruthenium catalyst, HCTU, DIEA, piperidine, sodium hexamethyldisilylamide, lithium hexamethyldisilylamide and Ac2O were purchased from Aldrich (Germany), and TFA from Fluka (Germany). Peptide Ac-PEI(EQSVK)IS-Nle-K-Nle-NH2 (11) cyclized through an amide bond between E4 and K8 was purchased from Peptide Protein Research (UK). The pure product was analyzed by HPLC and electrospray ionization mass spectrometry (ESI-MS). Fmoc-protected amino acids were purchased from Fluka, Novabiochem (Merck, Germany) and Iris Biotech (Germany). Fmoc-protected Rink Amide MBHA resin (0.47 mmol g−1 loading) was purchased from GS Biochem (China). All amino acids used were of the L-configuration. Linear precursor peptides 12 and 13 were synthesized manually on a 20-positions manifold (Omega) in a 20 mL polypropylene syringe (Dubelcco) equipped with a porous polyethylene filter following SPPS protocols. The coupling and RCM reactions were carried out on solid phase using microwave radiation in a Biotage Initiator reactor in a 5 mL vial. The rest of the SPPS reactions were stirred using an IKA-100 orbital shaker. After cleavage, the acidic crudes were sedimented in Et2O on a Hettlich Universal 320R centrifuge at 5000 rpm. All the crude and samples were lyophilized using mixtures H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH3CN on a Telstar 8-80 instrument. The monitoring of the reactions was also performed by HPLC/MS through a Waters 12695 HPLC connected to a Waters Micromass ZQ spectrometer. Peptides were purified by reverse-phase flash chromatography using C-18 derivatized silica cartridges (C18 Discovery SPE, Aldrich) in a manifold system or by semipreparative HPLC on a Waters 600 apparatus using an ACE 5 C18-300 (10 × 250 mm) column at a flow rate of 6 mL min−1 and a Waters 2487 detector, monitored at 214 nm. Mixtures of CH3CN (solvent A) and H2O with 0.05% TFA (solvent B) were used a mobile phase. The purity of the synthesized peptides was checked by an HPLC system (Waters 600 or an Agilent equipment) using in all cases a Sunfire C18 column (4.6 mm × 150 mm, 3.5 μm) and the same solvents as the mobile phase (1.0 mL min−1 flow). Peak detection was carried out at 214 and 254 nm. High-resolution mass spectra (HRMS) were obtained in an Agilent 6520 Accurate-Mass Q-TOF LC/MS mass spectrometer using 1% formic acid MeOH or MeOH–CH3CN mixtures. 1H and 13C-NMR spectra were recorded in general in CDCl3, acetone-d6 or DMSO-d6 on a Varian INOVA-400 (400 & 100 MHz), a Varian MERCURY-400 (400 & 100 MHz), a Varian UNITY-500 (500 & 125 MHz) or in a Bruker Avance 600 (600.13 & 150.03 MHz) spectrometer using TMS or DSS as the internal reference. The latter was equipped with a TXI cryoprobe. NMR experiments of helix-constrained analogues 5a, 8a and 11 were recorded at 1–2 mM concentration in 30% TFE/H2O solution. 1H- and 13C-NMR chemical shifts (δ) assignation of the former peptides was performed following the standard sequential assignation method using the SPARKY software.36 Optical rotation measurements were performed in a Perkin Elmer 241 MC using a 1 mL cell at 589 nm (sodium D-line).
CH3CN on a Telstar 8-80 instrument. The monitoring of the reactions was also performed by HPLC/MS through a Waters 12695 HPLC connected to a Waters Micromass ZQ spectrometer. Peptides were purified by reverse-phase flash chromatography using C-18 derivatized silica cartridges (C18 Discovery SPE, Aldrich) in a manifold system or by semipreparative HPLC on a Waters 600 apparatus using an ACE 5 C18-300 (10 × 250 mm) column at a flow rate of 6 mL min−1 and a Waters 2487 detector, monitored at 214 nm. Mixtures of CH3CN (solvent A) and H2O with 0.05% TFA (solvent B) were used a mobile phase. The purity of the synthesized peptides was checked by an HPLC system (Waters 600 or an Agilent equipment) using in all cases a Sunfire C18 column (4.6 mm × 150 mm, 3.5 μm) and the same solvents as the mobile phase (1.0 mL min−1 flow). Peak detection was carried out at 214 and 254 nm. High-resolution mass spectra (HRMS) were obtained in an Agilent 6520 Accurate-Mass Q-TOF LC/MS mass spectrometer using 1% formic acid MeOH or MeOH–CH3CN mixtures. 1H and 13C-NMR spectra were recorded in general in CDCl3, acetone-d6 or DMSO-d6 on a Varian INOVA-400 (400 & 100 MHz), a Varian MERCURY-400 (400 & 100 MHz), a Varian UNITY-500 (500 & 125 MHz) or in a Bruker Avance 600 (600.13 & 150.03 MHz) spectrometer using TMS or DSS as the internal reference. The latter was equipped with a TXI cryoprobe. NMR experiments of helix-constrained analogues 5a, 8a and 11 were recorded at 1–2 mM concentration in 30% TFE/H2O solution. 1H- and 13C-NMR chemical shifts (δ) assignation of the former peptides was performed following the standard sequential assignation method using the SPARKY software.36 Optical rotation measurements were performed in a Perkin Elmer 241 MC using a 1 mL cell at 589 nm (sodium D-line).
Synthetic procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DBU
DBU![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMF, 1
DMF, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 (1 × 1 min) and (3 × 10 min) (incomplete Fmoc deprotection was observed with the standard 20% piperidine in DMF) and the resin was drained and washed with DMF/CH2Cl2/DMF/CH2Cl2/DMF (4 × 0.5 min). Then, a solution of the corresponding N-Fmoc-protected amino acid (1.2 equiv), HCTU (1.2 equiv) and DIEA (2.4 equiv) in anhydrous DMF (0.5–1.0 mL) was added over the swollen peptidil-resin (1.0 equiv) in anhydrous DMF in a microwave vial (5–10 mL). The coupling reaction was heated at 40 °C using microwave radiation for 10 min. Each coupling step was repeated 3 times using freshly prepared Fmoc-amino acid/coupling reagent mixtures. After complete couplings, the resin was drained and washed with DMF/CH2Cl2/DMF/CH2Cl2 (4 × 0.5 min). Coupling reactions to primary and secondary amines were monitored by the ninhydrin and the chloranil tests, respectively.
48 (1 × 1 min) and (3 × 10 min) (incomplete Fmoc deprotection was observed with the standard 20% piperidine in DMF) and the resin was drained and washed with DMF/CH2Cl2/DMF/CH2Cl2/DMF (4 × 0.5 min). Then, a solution of the corresponding N-Fmoc-protected amino acid (1.2 equiv), HCTU (1.2 equiv) and DIEA (2.4 equiv) in anhydrous DMF (0.5–1.0 mL) was added over the swollen peptidil-resin (1.0 equiv) in anhydrous DMF in a microwave vial (5–10 mL). The coupling reaction was heated at 40 °C using microwave radiation for 10 min. Each coupling step was repeated 3 times using freshly prepared Fmoc-amino acid/coupling reagent mixtures. After complete couplings, the resin was drained and washed with DMF/CH2Cl2/DMF/CH2Cl2 (4 × 0.5 min). Coupling reactions to primary and secondary amines were monitored by the ninhydrin and the chloranil tests, respectively.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added (1 × 1 min) and (4 × 10 min) and washed with DMF/CH2Cl2/DMF/CH2Cl2/DMF (4 × 0.5 min). The cyclic resin-bounded peptides were then cleaved from the resin by treatment with an acidolitic mixture of TFA
1) was added (1 × 1 min) and (4 × 10 min) and washed with DMF/CH2Cl2/DMF/CH2Cl2/DMF (4 × 0.5 min). The cyclic resin-bounded peptides were then cleaved from the resin by treatment with an acidolitic mixture of TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TIPS
TIPS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 90
H2O, 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (6 mL) for 4 h at room temperature. The filtrates were precipitated from cold diethylether (50 mL), centrifuged at 5000 rpm (3 × 10 min) and lyophilized. The cleaved products were purified by reverse phase chromatography using SPE cartridges or semipreparative HPLC.
5 (6 mL) for 4 h at room temperature. The filtrates were precipitated from cold diethylether (50 mL), centrifuged at 5000 rpm (3 × 10 min) and lyophilized. The cleaved products were purified by reverse phase chromatography using SPE cartridges or semipreparative HPLC.
Ac-Pro-Glu-Ile-cycle[(CH2)6CH
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/i_char_e001.gif) CH(CH2)3]4,11[(D)-Ala-Gln-Ser-Val-Gly-Ile-Ser-Ala]-Lys-Nle-NH2 (5a, b). After elongation of the peptide starting from 0.05 mmol of Rink amide resin, the metathesis reaction was carried out for 30 min following the general protocol under microwave conditions. Then, the cyclic peptide was N-acetylated and cleaved from the resin. The crude was purified by reverse phase chromatography using SPE cartridges to give the cyclic peptide 5a, b (23.7 mg, 30% overall yield) as a partially solved mixture of E/Z isomers (85/15) in three fractions: major isomer (12.0 mg), minor isomer (1.0 mg) and mixture (10.7 mg). HPLC (waters, 5% to 100% of water in 30 min): 4.06 min (5a, major isomer, 92% analytical purity) and 3.89 min (5b, minor isomer). Major isomer 5a: HRMS (ESI, +) m/z: calculated for C71H122N16O19 1502.9017; found 752.4580 [(M + 2H)/2]+. 1H-NMR (600 MHz, 30% TFA D2O
CH(CH2)3]4,11[(D)-Ala-Gln-Ser-Val-Gly-Ile-Ser-Ala]-Lys-Nle-NH2 (5a, b). After elongation of the peptide starting from 0.05 mmol of Rink amide resin, the metathesis reaction was carried out for 30 min following the general protocol under microwave conditions. Then, the cyclic peptide was N-acetylated and cleaved from the resin. The crude was purified by reverse phase chromatography using SPE cartridges to give the cyclic peptide 5a, b (23.7 mg, 30% overall yield) as a partially solved mixture of E/Z isomers (85/15) in three fractions: major isomer (12.0 mg), minor isomer (1.0 mg) and mixture (10.7 mg). HPLC (waters, 5% to 100% of water in 30 min): 4.06 min (5a, major isomer, 92% analytical purity) and 3.89 min (5b, minor isomer). Major isomer 5a: HRMS (ESI, +) m/z: calculated for C71H122N16O19 1502.9017; found 752.4580 [(M + 2H)/2]+. 1H-NMR (600 MHz, 30% TFA D2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 1
H2O, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) and 13C-NMR (150 MHz, 30% TFA D2O
9) and 13C-NMR (150 MHz, 30% TFA D2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 1
H2O, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) in Table S3 (ESI†).
9) in Table S3 (ESI†).
Ac-Pro-Glu-Ile-cycle[(CH2)4CH
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/i_char_e001.gif) CH(CH2)4]4,8[(D)-Ala-Gln-Ser-Val-Ala]-Ile-Ser-Nle-Lys-Nle-NH2 (8a, b). After elongation of the peptide starting from 0.07 mmol of Rink amide resin, the metathesis reaction was carried out for 150 min following the general protocol under microwave conditions. After N-acetylation and cleavage from the resin, the crude was purified by semipreparative HPLC (waters, 3% to 30% of water, linear gradient in 27 min) to yield the cyclic derivative 8a, b (7.8 mg, 6% overall yield) as a partially solved mixture of E/Z isomers (60
CH(CH2)4]4,8[(D)-Ala-Gln-Ser-Val-Ala]-Ile-Ser-Nle-Lys-Nle-NH2 (8a, b). After elongation of the peptide starting from 0.07 mmol of Rink amide resin, the metathesis reaction was carried out for 150 min following the general protocol under microwave conditions. After N-acetylation and cleavage from the resin, the crude was purified by semipreparative HPLC (waters, 3% to 30% of water, linear gradient in 27 min) to yield the cyclic derivative 8a, b (7.8 mg, 6% overall yield) as a partially solved mixture of E/Z isomers (60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40) in two fractions: major isomer (2.1 mg) and mixture (5.7 mg). HPLC (Agilent, 10% to 100% of water in 10 min): 6.68 min (8a, major isomer, 90% analytical purity) and 6.49 min (8b, minor isomer). Major isomer 8a: HRMS (ESI, +) m/z: calculated for C72H124N16O19 1516.9208; found 759.4669 [(M + 2H)/2]+. 1H-NMR (600 MHz, 30% TFA D2O
40) in two fractions: major isomer (2.1 mg) and mixture (5.7 mg). HPLC (Agilent, 10% to 100% of water in 10 min): 6.68 min (8a, major isomer, 90% analytical purity) and 6.49 min (8b, minor isomer). Major isomer 8a: HRMS (ESI, +) m/z: calculated for C72H124N16O19 1516.9208; found 759.4669 [(M + 2H)/2]+. 1H-NMR (600 MHz, 30% TFA D2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 1
H2O, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) and 13C-NMR (150 MHz, 30% TFA D2O
9) and 13C-NMR (150 MHz, 30% TFA D2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 1
H2O, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) in Table S4 (ESI†).
9) in Table S4 (ESI†).
Ac-Pro-Glu-Ile-cycle[(CH2)4CONH(CH2)4]4,8[Glu-Gln-Ser-Val-Lys]-Ile-Ser-Nle-Lys-Nle-NH2 (11). This peptide, cyclized through an amide bond between E4 and K8, was purchased from Peptide Protein Research (UK). The pure product was analyzed by HPLC and electrospray ionization mass spectrometry (ESI-MS). HPLC (Agilent, 10% to 100% of water in 10 min): 5.98 min, 95% analytical purity. HRMS (ESI, +) m/z: calculated for C69H119N17O20 1505.8823; found 1506.8900 [M + H]+. 1H-NMR (600 MHz, 30% TFA D2O
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 1
H2O, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) and 13C-NMR (150 MHz, 30% TFA D2O
9) and 13C-NMR (150 MHz, 30% TFA D2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 1
H2O, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9) in Table S5 (ESI†).
9) in Table S5 (ESI†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (100 μL). 30 μL of the resulting quenched reaction was injected onto an HPLC-MS (through a HPLC-waters 12695 connected to a Waters Micromass ZQ spectrometer), and the amount of starting peptide present was quantified by mass integration of the peak at 214 nm. Duplicate or triplicate reactions were run for each time point reported and half-lives were determined by fitting time dependent peptide concentration to an exponential decay using GraphPad Prism.
1 (100 μL). 30 μL of the resulting quenched reaction was injected onto an HPLC-MS (through a HPLC-waters 12695 connected to a Waters Micromass ZQ spectrometer), and the amount of starting peptide present was quantified by mass integration of the peak at 214 nm. Duplicate or triplicate reactions were run for each time point reported and half-lives were determined by fitting time dependent peptide concentration to an exponential decay using GraphPad Prism.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 30
1 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 63
63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 in volume at pH 5.5. This pH was selected because amide NH signals are more intense and sharper at pH 5.5 than at pH 7.0, and the pKa values for the charged side chains in these peptides (Glu & Lys) lie out of this pH range. Hence, peptide conformation will be the same within this pH range. 1D 1H spectra, 2D homonuclear (1H,1H-COSY, 60 ms 1H,1H-TOCSY, and 150 ms 1H,1H-NOESY) spectra, and 2D heteronuclear 1H,13C-HSQC spectra at natural 13C abundance were acquired as previously described38 using a Bruker AV-600 spectrometer operating at a 1H frequency of 600.13 MHz and equipped with a cryoprobe. Data were processed using the TOPSPIN program.39 1H- and 13C-NMR chemical shifts (δ) were assigned following the standard sequential assignment method40 using the SPARKY software.36
7 in volume at pH 5.5. This pH was selected because amide NH signals are more intense and sharper at pH 5.5 than at pH 7.0, and the pKa values for the charged side chains in these peptides (Glu & Lys) lie out of this pH range. Hence, peptide conformation will be the same within this pH range. 1D 1H spectra, 2D homonuclear (1H,1H-COSY, 60 ms 1H,1H-TOCSY, and 150 ms 1H,1H-NOESY) spectra, and 2D heteronuclear 1H,13C-HSQC spectra at natural 13C abundance were acquired as previously described38 using a Bruker AV-600 spectrometer operating at a 1H frequency of 600.13 MHz and equipped with a cryoprobe. Data were processed using the TOPSPIN program.39 1H- and 13C-NMR chemical shifts (δ) were assigned following the standard sequential assignment method40 using the SPARKY software.36Distance constraints for structure calculation were derived from 2D NOESY spectra. The automatic integration subroutine of the Sparky program36 was used to integrate the NOE cross-peaks. Dihedral angle restraints for ϕ and ψ angles were derived from 1H, 13Cα and 13Cβ chemical shifts using the TALOS program.41 Since TALOS does not include non-natural residues, Nle and the hydrocarbon staple's residues were considered as Lys for this program. Structure calculations were performed using the program CYANA 2.1.42 First, the non-natural residue Nle and those required for the covalent closure in peptide 8a (amino acid (S)-3) and 11 (Glu and Lys of the amide bridge) were generated in the format required for the CYANA library. Then, the standard iterative protocol for automated NOE assignment was run using as input the list of integrated NOE cross-peaks and the full list of chemical shifts, and as additional restraints, the TALOS-derived ϕ, ψ dihedral angle ranges, and the upper and lower limit restraints required to link residues at positions 4 and 8. The iterative protocol consists of seven cycles of combined automated NOE assignment and structure calculation of 100 conformers per cycle, and a final step of standard annealing calculation of 100 conformers.43 The ensemble of the 20 lowest target function conformers resulting from were examined and visualised with MOLMOL44 and PyMOL.
To build the peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Li-TryR complexes, the corresponding peptide was overlaid onto the Pro435-Met447 α-helix of monomer B in the crystal structure of the enzyme. Then, a 30 ns unrestrained MD simulation was performed for each system following the general protocol. The total and per-residue peptide–protein binding energies (kcal mol−1) were calculated by means of the fast and versatile program MM-ISMSA47 using the snapshots collected during the last 20 ns of the respective MD simulations.
Li-TryR complexes, the corresponding peptide was overlaid onto the Pro435-Met447 α-helix of monomer B in the crystal structure of the enzyme. Then, a 30 ns unrestrained MD simulation was performed for each system following the general protocol. The total and per-residue peptide–protein binding energies (kcal mol−1) were calculated by means of the fast and versatile program MM-ISMSA47 using the snapshots collected during the last 20 ns of the respective MD simulations.
Conflict of interest
The authors declare no financial or commercial conflict of interest.Abbreviations
| DSSP | Define secondary structure of proteins algorithm |
| ELISA | Enzyme-Linked ImmunoSorbent Assay |
| Li-TryR | Leishmania infantum trypanothione reductase |
| MD | Molecular dynamics |
| RCM | Ring-closing metathesis |
| RMSD | Root mean square deviation |
| SPE | Solid-phase extraction |
| SPPS | Solid phase peptide synthesis. |
Acknowledgements
PASM, MAT and HL thank the Spanish MEC/MICINN for their FPI fellowships. Financial support from the Spanish MINECO (Projects SAF2012-39760-C02 and CTQ2011-22514), Comunidad de Madrid (S-2010/BMD-2457; BIPEDD-2) and Junta de Comunidades de Castilla la Mancha (Project POII10-0180-7897) is gratefully acknowledged.Notes and references
- WHO, Wold Health Organization, 2015, available in http://www.who.int/topics/lesihmaniasis/en/, accessed May 2015.
- (a) N. Singh, M. Kumar and R. K. Singh, Asian Pac. J. Trop. Med., 2012, 5, 485 CrossRef CAS PubMed; (b) H. Hussain, A. Al-Harrasi, A. Al-Rawahi, I. R. Green and S. Gibbons, Chem. Rev., 2014, 114, 10369 CrossRef CAS PubMed.
- M. Zucca, S. Scutera and D. Savoia, Curr. Med. Chem., 2013, 20, 502 CAS.
- K. Ait-Oudhia, E. Gazanion, B. Vergnes, B. Oury and D. Sereno, Parasitol. Res., 2011, 109, 1225 CrossRef PubMed.
- T. T. H. Pham, P. M. Loiseau and G. Barratt, Int. J. Pharm., 2013, 454, 539 CrossRef CAS PubMed.
- A. H. Fairlamb, P. Blackburn, P. Ulrich, B. T. Chait and A. Cerami, Science, 1985, 227, 1485 CAS.
- R. L. Krauth-Siegel, H. Bauer and R. H. Schirmer, Angew. Chem., Int. Ed., 2005, 44, 690 CrossRef CAS PubMed.
- See for example, L. S. C. Bernades, C. L. Zani and I. Carvalho, Curr. Med. Chem., 2013, 20, 2673 CrossRef , and references therein.
- See for example: (a) T. J. Benson, J. H. Mckie, J. Garforth, A. Borges, A. H. Fairlamb and K. T. Douglas, Biochem. J., 1992, 286, 9 CrossRef CAS PubMed; (b) C. Chan, H. Yin, J. Garforth, J. H. McKie, R. Jaouhari, P. Speers, K. T. Douglas, P. J. Rock, V. Yardley, S. L. Croft and A. H. Fairlamb, J. Med. Chem., 1998, 41, 148 CrossRef CAS PubMed; (c) M. O. F. Khan, S. E. Austin, C. Chan, H. Yin, D. Marks, S. N. Vaghjiani, H. Kendrick, V. Yardley, S. L. Croft and K. T. Douglas, J. Med. Chem., 2000, 43, 3148 CrossRef CAS PubMed.
- See also for example: (a) J. L. Richardson, I. R. E. Nett, D. C. Jones, M. H. Abdile, I. H. Gilbert, A. H. Fairlamb, S. Patterson, D. C. Jones, E. J. Shanks, J. A. Frearson and P. G. Wyatt, ChemMedChem, 2009, 4, 1333 CrossRef CAS PubMed; (b) S. K. Venkatesan and D. V. K. Dubey, Sci. World J., 2012, 1 CrossRef PubMed.
- See for example: (a) M. C. O'Sullivan, Q. B. Zhou, Z. Li, T. B. Durham, D. Rattendi, S. Lane and C. J. Bacchi, Bioorg. Med. Chem., 1997, 5, 2145–2155 CrossRef; (b) B. Bonnet, D. Soullez, S. Girault, L. Maes, V. Landry, E. Davioud-Charvet and C. Sergheraert, Bioorg. Med. Chem., 2000, 8, 95 CrossRef CAS PubMed.
- (a) M. R. Ariyanayagam, S. L. Oza, A. Mehlert and A. H. Fairlamb, J. Biol. Chem., 2003, 278, 27612 CrossRef CAS PubMed; (b) M. J. Dixon, R. I. Maurer, C. Biggi, J. Oyarzabal, J. W. Essex and M. Bradley, Bioorg. Med. Chem., 2005, 13, 4513 CrossRef CAS PubMed.
- (a) A. Tromelin, M. Moutiez, D. Meziane-Cherif, M. Aumercier, A. Tartar and C. Sergheraert, Bioorg. Med. Chem. Lett., 1993, 3, 1971 CrossRef CAS; (b) E. A. Garrard, E. C. Borman, B. N. Cook, E. J. Pike and D. G. Alberg, Org. Lett., 2000, 2, 3639 CrossRef CAS PubMed.
- (a) S. Patterson, M. S. Alphey, D. C. Jones, E. J. Shanks, I. P. Street, J. A. Frearson, P. G. Wyatt, I. H. Gilbert and A. H. Fairlamb, J. Med. Chem., 2011, 54, 6514 CrossRef CAS PubMed; (b) M. H. Duyzend, C. T. Clark, S. L. Simmons, W. B. Johnson, A. M. Larson, A. M. Leconte, A. W. Wills, M. Ginder-Vogel, A. K. Wihelm, J. A. Czechowicz and D. G. Alberg, J. Enzyme Inhib. Med. Chem., 2012, 27, 784 CrossRef CAS PubMed.
- M. A. Toro, P. A. Sánchez-Murcia, D. Moreno, M. Ruiz-Santaquiteria, J. F. Alzate, A. Negri, M. J. Camarasa, F. Gago, S. Velázquez and A. Jiménez-Ruiz, ChemBioChem, 2013, 14, 1212 CrossRef CAS PubMed.
- See for example reviews: (a) L. K. Henchey, A. L. Jochim and P. S. Arora, Curr. Opin. Chem. Biol., 2008, 12, 692 CrossRef CAS PubMed; (b) A. Grauer and B. König, Eur. J. Org. Chem., 2009, 5099 CrossRef CAS; (c) R. M. J. Liskamp, D. T. S. Rijkers, J. A. W. Kruijtzer and J. Kemmink, ChemBioChem, 2011, 12, 1626 CrossRef CAS PubMed; (d) V. Azzarito, K. Long, N. S. Murphy and A. J. Wilson, Nat. Chem., 2013, 5, 161 CrossRef CAS PubMed; (e) T. A. Hill, N. E. Shepherd, F. Diness and D. P. Fairlie, Angew. Chem., Int. Ed., 2014, 53, 13020 CrossRef CAS PubMed.
- See for example review: J. W. Taylor, Biopolymers, 2002, 66, 49 CrossRef CAS PubMed.
- See for example: (a) J. C. Phelan, N. J. Skelton, A. C. Braidsted and R. S. McDowell, J. Am. Chem. Soc., 1997, 119, 455 CrossRef CAS; (b) S. G. Yao, M. A. Smith-White, E. K. Potter and R. S. Norton, J. Med. Chem., 2002, 45, 2310 CrossRef CAS PubMed; (c) N. E. Shepherd, H. N. Hoang, V. S. Desai, E. Letoize, P. R. Young and D. P. Fairlie, J. Am. Chem. Soc., 2006, 128, 13284 CrossRef CAS PubMed.
- See for example: (a) M. I. García-Aranda, Y. Mirassou, B. Gautier, M. Martín-Martínez, N. Inguimbert, M. Vidal, M. T. García-López, M. A. Jiménez, R. González-Muñiz and M. J. Pérez de Vega, Bioorg. Med. Chem., 2011, 19, 7526 CrossRef PubMed; (b) K. K. Khoo, M. J. Wilson, B. J. Smith, M. Zhang, J. Gulyas, D. Yoshikami, J. E. Rivier, G. Bulaj and R. S. Norton, J. Med. Chem., 2011, 54, 7558 CrossRef CAS PubMed; (c) M. I. García-Aranda, S. González-López, C. M. Santiveri, N. Gagey-Eilstein, M. Reille-Seroussi, M. Martín-Martínez, N. Inguimbert, M. Vidal, M. T. García-López, M. A. Jiménez, R. González-Muñiz and M. J. Pérez de Vega, Org. Biomol. Chem., 2013, 11, 1896 RSC.
- See recent reviews: (a) A. Brik, Adv. Synth. Catal., 2008, 350, 1661 CrossRef CAS; (b) M. J. Pérez de Vega, M. I. García-Aranda and R. González-Muñiz, Med. Res. Rev., 2011, 31, 677 Search PubMed; (c) G. L. Verdine and G. J. Hilinski, Methods Enzymol., 2012, 503, 3–33 CAS; (d) R. Dharanipragada, Future Med. Chem., 2013, 5, 831 CrossRef CAS PubMed; (e) L. D. Walensky and G. H. Bird, J. Med. Chem., 2014, 57, 6275 CrossRef CAS PubMed.
- See for example: (a) C. E. Schafmeister, J. Po and G. L. Verdine, J. Am. Chem. Soc., 2000, 122, 5891 CrossRef CAS; (b) H. E. Blackwell and R. H. Grubbs, Angew. Chem., Int. Ed., 1998, 37, 3281 CrossRef CAS; (c) P. S. Kutchukian, J. S. Yang, G. L. Verdine and E. I. Shakhnovich, J. Am. Chem. Soc., 2009, 131, 4622 CrossRef CAS PubMed.
- (a) F. Bernal, A. F. Tyler, S. J. Korsmeyer, L. D. Walensky and G. L. Verdine, J. Am. Chem. Soc., 2007, 129, 2456 CrossRef CAS PubMed; (b) F. Bernal, M. Wade, M. Godes, T. N. Davis, D. G. Whitehead, A. L. Kung, G. M. Wahl and L. D. Walensky, Cancer Cell, 2010, 411 CrossRef CAS PubMed; (c) L. D. Walensky, A. L. Kung, I. Escher, T. J. Malia, S. Barbuto, R. Wright, G. Wagner, G. L. Verdine and S. J. Korsmeyer, Science, 2004, 305, 1466 CrossRef CAS PubMed; (d) L. D. Walensky, K. Pitter, J. Morash, K. J. Oh, S. Barbuto, J. Fisher, E. Smith, G. L. Verdine and S. J. Korsmeyer, Mol. Cell, 2006, 24, 199 CrossRef CAS PubMed; (e) N. N. Danial, L. D. Walensky and C.-Y. Zhang, et al., Nat. Med., 2008, 14, 144 CrossRef CAS PubMed.
- (a) R. E. Moellering, M. Cornejo, T. N. Davis, C. Del Bianco, J. C. Aster, S. C. Blacklow, A. L. Kung, D. G. Gilliland, G. L. Verdine and J. E. Bradner, Nature, 2009, 462, 182 CrossRef CAS PubMed; (b) G. H. Bird, N. Madani, A. F. Perry, A. M. Princiotto, J. G. Supko, X. He, E. Gavathiotis, J. G. Sodroski and L. D. Walensky, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14093 CrossRef CAS PubMed; (c) T.-L. Sun, Y. Sun, C.-C. Lee and H. W. Huang, Biophys. J., 2013, 104, 1923 CrossRef CAS PubMed; (d) Y.-Q. Long, S.-X. Huang and Z. Zawahir, J. Med. Chem., 2013, 56, 5601 CrossRef CAS PubMed; (e) H.-K. Cui, J. Qing, Y. Guo, Y.-J. Wang, L. J. Cui, T.-H. He, L. Zhang and L. Liu, Bioorg. Med. Chem., 2013, 21, 3547 CrossRef CAS PubMed.
- The nomenclature Si, i+4S(8) refers to a 8-carbon tether with S-configuration at both i, i + 4 positions.
- The nomenclature Ri, i+7S(11) refers to a 11-carbon tether with R-configuration at i, i + 4 position and S-configuration at i, i + 7 position.
- Y. W. Kim and G. L. Verdine, Bioorg. Med. Chem., 2009, 19, 2533 CrossRef CAS PubMed.
- R. M. Williams and M. N. Im, J. Am. Chem. Soc., 1991, 113, 9276 CrossRef CAS.
- A. J. Robinson, J. Elaridi, B. J. Van Lierop, S. Mujcinovic and W. R. Jackson, J. Pep. Sci., 2007, 13, 280 CrossRef CAS PubMed.
- The spectra of peptides 8a and 11 could not be carried out in absence of TFE due to poor water-solubility of the peptides. It should be noted that TFE is not a general α-helix-inducer, as generally believed (see reference: M. Buck, Q. Rev. Biophys., 1998, 31, 297). Indeed, it has been shown to stabilise non-helical structures, such as β-hairpins (see reference: C. M. Santiveri, D. Pantoja-Uceda, M. Rico and M. A. Jiménez, Biopolymers, 2005, 79150).
- (a) R. Vila, I. Ponte, P. Suau, M. A. Jiménez and M. Rico, Protein Sci., 2000, 9, 627 CrossRef CAS PubMed; (b) M. A. Jiménez, A. C. Barrachi-Saccilotto, E. Valdivia, M. Maqueda and M. Rico, J. Pept. Sci., 2005, 11, 29 CrossRef PubMed.
- P. Güntert, Automated NMR protein structure calculation with CYANA, Methods Mol. Biol., 2004, 278, 353 Search PubMed.
- T. Okamoto, K. Zobel, A. Fedorova, C. Quan, H. Yang, W. J. Fairbrother, D. C. S. Huang, B. J. Smith, K. Deshayes and P. E. Czabotar, ACS Chem. Biol., 2013, 8, 297 CrossRef CAS PubMed.
- D. J. Derksen, J. L. Stymiest and J. C. Vederas, J. Am. Chem. Soc., 2006, 128, 14252 CrossRef CAS PubMed.
- During the proteolytic stability studies, opening of the lactam-bridge of analogue 11 was not detected by HPLC-MS. This points to the stability of the amide bridge under these conditions.
- See for example: (a) N. E. Shepherd, G. Abbenante and D. P. Fairlie, Angew. Chem., Int. Ed., 2004, 43, 2687 CrossRef CAS; (b) E. N. Murgae, G. Gao, A. Bisello and J. Ahn, J. Med. Chem., 2010, 53, 6421 CrossRef PubMed; (c) T. Rao, G. Ruiz-Gomez, T. A. Hill, H. N. Hoang, D. P. Fairlie and J. M. Mason, PLoS One, 2013, 8, e59415 Search PubMed.
- T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco Search PubMed.
- (a) J. L. Stymiest, B. F. Mitchell, S. Wong and J. C. Vederas, Org. Lett., 2003, 5, 47 CrossRef CAS PubMed; (b) J. L. Stymiest, B. F. Mitchell, S. Wong and J. C. Vederas, J. Org. Chem., 2005, 70, 7799 CrossRef CAS PubMed.
- C. M. Santiveri, D. Pantoja-Uceda, M. Rico and M. A. Jiménez, Biopolymers, 2005, 79, 150 CrossRef CAS PubMed.
- NMR data and processing software, Bruker Biospin, Kalsruhe, Germany Search PubMed.
- K. Wüthrich, NMR of Proteins and Nucleic acids, John Wiley & Sons, New York, 1986 Search PubMed.
- G. Cornilescu, F. Delaglio and A. Bax, J. Biomol. NMR, 1999, 13, 289 CrossRef CAS PubMed.
- P. Guntert, C. Mumemthaler and K. Wutrich, J. Mol. Biol., 1997, 273, 283 CrossRef CAS PubMed.
- P. Guntert, Prog. Nucl. Magn. Reson. Spectrosc., 2004, 43, 105 CrossRef.
- R. Koradi, M. Billeter and K. Wuthrich, J. Mol. Graphics, 1996, 14, 51 CrossRef CAS PubMed.
- D. A. Case, T. A. Darden, T. E. Cheatham, C. L. Simmerling, J. Wang, R. E. Duke, R. Luo, R. C. Walker, W. Zhang, K. M. Merz, B. Roberts, S. Hayik, A. Roitberg, G. Seabra, J. Swails, A. W. Goetz, I. Kolossvary, K. F. Wong, F. Paesani, J. Vanicek, R. M. Wolf, J. Liu, X. Wu, S. R. Brozell, T. Steinbrecher, H. Gohlke, Q. Cai, X. Ye, J. Wang, M. J. Hsieh, G. Cui, D. R. Roe, D. H. Mathews, M. G. Seetin, R. Salomon-Ferrer, C. Sagui, V. Babin, T. Luchko, S. Gusarov, A. Kovalenko and P. A. Kollman, Amber 12, 2012 Search PubMed.
- C. Sander and W. Kabsch, Biopolymers, 1983, 22, 2577 CrossRef PubMed.
- J. Klett, A. Núñez-Salgado, H. G. Dos Santos, A. Cortés-Cabrera, A. Perona, R. Gil-Redondo, D. Abia, F. Gago and A. Morreale, J. Chem. Theory Comput., 2012, 8, 3395 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra06853c |
| This journal is © The Royal Society of Chemistry 2015 |