Metabolic pathways in cancers: key targets and implications in cancer therapy
Sathya Narayanan Vijayakumar,
Swaminathan Sethuraman and
Uma Maheswari Krishnan*
Centre for Nanotechnology & Advanced Biomaterials, School of Chemical & Biotechnology, SASTRA University, Thanjavur-613401, Tamil Nadu, India. E-mail: umakrishnan@sastra.edu; Fax: +91 4362 264120; Tel: +91 4362 264101 ext. 3677
First published on 30th April 2015
Abstract
Proliferation and self-sufficiency are two of the most important properties of cancer cells. Although genetic aberrations are believed to be the reason for cancer development, the importance of metabolic alterations in cancer development has been in the lime light lately. The most challenging aspect in cancer treatment has been the similarity to host cells. The discovery of various metabolic alterations that occur in cancers to attain and maintain a proliferative state has resulted in new information on the metabolic differences between normal and cancer cells. One such alteration is the establishment of the Warburg effect. This review elaborates on various changes that lead to the establishment of the Warburg effect in cancer cells and their consequences. Understanding the metabolic uniqueness of various cancers can aid in the identification of novel molecular targets leading to more efficient strategies in cancer treatment.
1. Introduction
Cancer cells due to their rapid proliferation require a vast amount of energy in a short time span. It is well documented that cancer cells generate a significantly higher percentage of their cellular energy (ATP) by lactic acid fermentation irrespective of the oxygen content unlike normal cells that employ mitochondrial oxidative phosphorylation for ATP generation.1 Mitochondrial oxidative phosphorylation is more energy efficient and produces 32 ATP molecules per glucose molecule consumed compared to lactic acid fermentation that generates only 2 ATP molecules per glucose molecule consumed. Warburg hypothesized that the glycolytic phenotype adopted by cancer cells might be due to irreversibly damaged mitochondria and cancer cells undergo a dormant or a ‘sleeping’ phase during which they develop significantly higher glycolytic ability through selective pressure to compensate the loss of mitochondrial function.1 Cells that fail to adopt and possess low glycolytic ability perish. While Warburg considered this glycolytic phenotype to be irreversible, Crabtree demonstrated the existence of a reversible switch between glycolytic phenotype and oxidative phosphorylation depending on glucose availability in some cancer cells.2 It has been found that even in the presence of completely functional mitochondria, lactic acid fermentation is not completely suppressed in normal cells. This reversible switch depending on glucose availability is termed as ‘Crabtree effect’. When Warburg reported his observation that cancerous cells depend on glycolysis for generation of ATP, the necessity of this adoptation in cancer development was poorly understood. Today, although mysteries of cancer are not completely solved, we have sufficient knowledge about signalling mechanisms involved and the consequences of Warburg effect that provide cancers cells proliferative advantage. The metabolic changes that facilitate the establishment of Warburg effect and its consequences in cancer proliferation and survival are discussed in this review. The implications of such changes in cancer therapy have also been discussed.2. Cancer energy metabolism
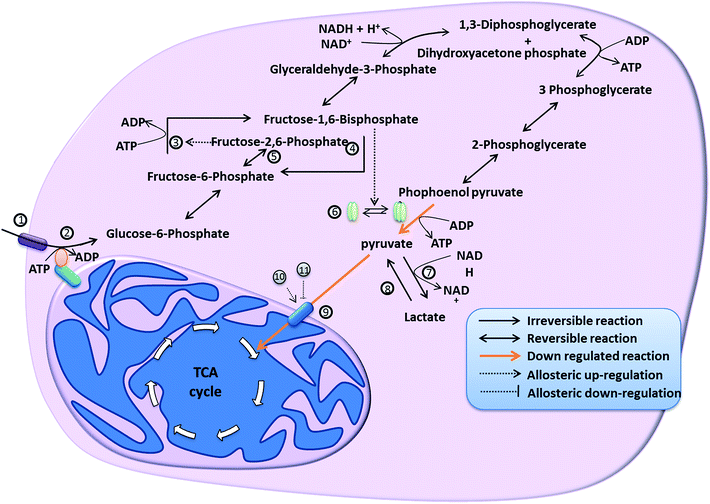
Uncontrolled proliferation is the primary property of most cancer cells.3 Rapid proliferation of cells requires high levels of ATP (energy), nucleotides, amino acids, lipids, etc. These small molecules required for building complex macromolecular structures like nucleic acids, proteins, cell membrane etc., are derived from various intermediates of the cellular energy metabolism. Hence, to sustain their high proliferation rate, cancer cells should modify their energy metabolism in order to satisfy their biosynthetic requirements. Three important changes in glucose metabolism are required to establish aerobic glycolysis. The prime requisite is to ensure adequate availability of the substrate (glucose).4 Secondly, modifications to overcome the tight regulation of glycolytic enzymes are required. Finally, for lactic acid production to occur, the conversion of pyruvate to acetyl-coA and its entry into mitochondria should be prevented. The following section elaborates the strategies used by cancer cells to establish Warburg effect and changes in tri-carboxylic acid cycle (TCA) of cancer cells. Fig. 1 depicts the various reactions involved in the glycolytic pathway in cancer cells.2.1. Substrate availability
Hatanaka (1974) proposed that the transport of glucose across the cell membrane is the first rate-limiting step for carbohydrate metabolism.4 Glucose is transported through cell membrane in an energy-independent manner facilitated by glucose transporters (GLUT). Among various isoforms of GLUT transporters, the high affinity GLUT1 transporters are over-expressed in several cancer cells. Studies using Rat1 fibroblasts in vitro have shown that hypoxic conditions and oncogenic expression of the GTPase H-ras can together stimulate expression of GLUT1 mRNA.5 H-ras is also capable of independently triggering GLUT1 mRNA over-expression. H-ras transformed cells were found to express higher levels of the hypoxia inducible factor, HIF1-α. Higher promoter activity was detected in HIF responsive fragment of GLUT1 promoter in H-ras transformed Rat1 cells than in normal Rat1 cells. Mutation in the HIF1 binding site caused reduction in H-ras mediated up-regulation of GLUT1 mRNA confirming the association between HIF1 and GLUT1.5 It has also been found that activation of PI3K/Akt pathway up-regulates the expression of GLUT1 transporters.6 The protein kinase Akt, which is widely regarded to contribute to Warburg effect, was found to regulate the expression and trafficking of the GLUT1 transporters on activation through phosphorylation by PI3K.7 As substrate availability plays a key role in proliferation and survival of cancer cells, glucose transporters can serve as interesting target in treatment of cancer cells. The localization of glucose transporters is controlled by intracellular regulatory mechanisms.8 Expression of glucose transporters also shows tissue specificity thus reflecting the physiological properties of each tissue.9 There are two approaches in targeting glucose transporters. One is to reduce the expression of GLUT proteins using anti-sense oligonucleotides against GLUT genes. This method has been proved to be successful in breast cancer.10 Another approach is to use drugs that can interfere with glucose transport like D-allose.112.2. Hexokinase (HK)
Increased HK activity has been detected in cancer cells with more than 50% of this activity found in the mitochondrial fraction.12,13 Growth rate of cancer cells is directly proportional to specific activity of HK associated with the mitochondria.12 It is also well known that proliferation rate of cancer is directly correlated to higher glycolytic capacity.14 Increase in glycolytic ability of cancer cells therefore, is at least partly due to this association. In an interesting experiment, addition of glucose to mitochondrial fraction of normal cells did not affect mitochondrial respiration whereas addition of glucose to mitochondrial fraction of cancer cells resulted in altered mitochondrial respiration suggesting direct coupling of glycolysis with mitochondrial ATP generation.13HK exists in two different molecular forms – soluble and particulate. Soluble form of HK is less active and sensitive to feedback inhibition by glucose-6-phosphate while the particulate form is more active and insensitive to this feedback regulation.15 The HK bound to mitochondrial membrane has been reported to resemble particulate form of HK and is less sensitive to inhibition by glucose-6-phosphate.13 Mitochondrial protein interacting with HK has been reported to be voltage-dependent anion channel (VDAC).16 The interaction between HK with VDAC provides HK a preferential access to the ATP produced from oxidative phosphorylation. It has been demonstrated that hexokinase-2 (HK2), which interacts with mitochondria is preferentially expressed in various cancers.14,17 HK2 interaction with mitochondrial membrane can protect it from proteolytic degradation. Reporter gene studies have shown that hypoxic conditions, glucose and insulin show positive effect on HK2 promoter activation.14 In addition to promoter activation, it has also been shown that gene duplication is also involved in increased expression of HK.14 Regulation of expression and properties of HK has been reviewed in detail elsewhere.18
Glucose phosphorylation activity and mitochondrial binding ability of HK are both important for its protective effect on cancer cells.19 Akt regulates the interaction between VDAC and HK by affecting the phosphorylation state of both VDAC and HK. Upon activation by PI3K, phosphorylated Akt (pAkt) is translocated to mitochondria, where it accumulates in the matrix, inner and outer membranes.20 pAkt phosphorylates two mitochondrial proteins namely glycogen synthase kinase-3β (GSK3β) and β subunit of ATP synthase. GSK3β inhibits HK-VDAC interaction by phosphorylation of VDAC. Phosphorylation of GSK3β by pAkt reduces its activity. Thus pAkt facilitates HK–VDAC interaction by negatively regulating GSK3β activity. HK–VDAC interaction can be facilitated by phosphorylation of HK by pAkt in a more direct manner. Hexokinase contains a consensus sequence for phosphorylation by pAkt whose phosphorylation enhances HK–VDAC interaction. Thus, activation of PI3K/Akt signalling leads to an increase in the mitochondrial association of HK.20 Investigations are underway to check the efficacy of hexokinase inhibitors like 2-deoxyglucose and 3-bromopyruvate in overcoming challenges posed by cancers. A much more specific approach might be to target HD-VDAC interaction that plays a major role in the ability of cancer cells to metabolize glucose at a high rate and suppression of apoptosis.
2.3. Phosphofructokinase (PFK)
Phosphofructokinase-1 (PFK1) is the most important enzyme in the regulation of glycolytic flux as the regulatory machineries involved sense the cellular energy levels. PFK1 catalyses the forward reaction (phosphorylation of fructose-6-phosphate to fructose-1,6-bisphosphate using an ATP molecule) of a substrate cycle, whose reverse reaction (formation of fructose-6-phosphate from fructose-1,6-bisphosphate) is catalysed by bisphosphatase (BP1).21 Both enzymes are allosterically regulated by fructose-2,6-bisphosphate, ATP, ADP and AMP. While ATP allosterically activates BP1 and deactivates PFK1, ADP and AMP activate PFK and deactivate BP1. Substrate cycles are very sensitive to changes in the concentration of allosteric regulators and thus PFK1/BP1 substrate cycle provides a very sensitive means to regulate glycolytic flux based cellular energy demands. Fructose-2,6-bisphosphate is considered to be the most effective allosteric activator of PFK1.22,23 Fructose-2,6-bisphosphate level is controlled by a bi-functional enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases (PFK-2/FBPase-2) that catalyses both the conversion of fructose-6-phosphate to fructose-2,6-bisphosphate using an ATP molecule as well as the hydrolysis of fructose-2,6-bisphosphate to fructose-6-phosphate. It is evident that as PFK-2/FBPase-2 utilizes ATP for phosphorylation of fructose-6-phosphate, it may serve to relieve the inhibitory effect of ATP on PFK1. PFKFB3 and PFKFB4 isoforms of PFK-2/FBPase-2 are components of HIF1 induced response to hypoxic conditions.24 PFKFB3 gene that contains oncogene-like regulatory element in its mRNA is over expressed in several aggressive cancer types and protein synthesis is induced by hypoxic conditions25,26 Myc oncogene up-regulates the expression of PFK1.27 Liver-type PFK is preferentially expressed in many cancer cells. PFK expressed in tumour cells and normal cells show different sensitivity towards allosteric regulators.28 It has been found that PFK from human glioma cells are less sensitive to inhibition by citrate but highly sensitive to activation by fructose-2,6-bisphosphate.28 Reaction catalysed by PFK is the most important regulatory point in glycolytic flux regulation making it an important target in cancer treatments that exploits uniqueness of cancer energy metabolism. Drugs that can influence the levels of the allosteric regulators provide an interesting way to target PFK activity. For example, inhibition of ATP dependent citrate lyase leads to increase in the citrate levels in cytoplasm due to increased accumulation of citrate.29 Citrate is a well-known inhibitor of PFK hence inhibition of citrate lyase leads to inhibition of PKF and leads to reduction in glycolytic rate. Higher activity of PFK-2/FBPase-2 is specific phenomenon found in cancer cells, thus making it a potential cancer specific target.2.4. Pyruvate kinase (PK)
Cancer cells express embryonic M2 splice isoform of pyruvate kinase (PKM2) and this change is necessary for establishment of Warburg effect.30 When M2 splice isoform is replaced with M1 isoform (PKM1), which is widely expressed in adult cells, there is a reversal of Warburg effect that is manifested through a reduction in the amount of lactate produced by the cancer cells as well as in their tumorigenicity.30 PK can exist either as highly active tetramer or less active dimer.31 Fructose-1,6-bisphosphate levels play a key role in regulating the equilibrium between tetrameric and dimeric forms. Higher levels of fructose-1,6-bisphosphate shifts the equilibrium towards formation of tetrameric form thus leading to an increase in the rate of pyruvate production while at lower levels of fructose-1,6-bisphosphate, PKM2 exists predominantly in the less active dimeric form leading to accumulation of glycolytic intermediates.31 Thus PKM2 provides an efficient a way to maintain a balance between energy generation and accumulation of glycolytic metabolites for biosynthesis. The tetrameric form of PKM2 has been reported to interact with various other glycolytic enzymes, which leads to ‘proximity effect’ i.e. preferential access to substrate.32 The equilibrium between tetrameric and dimeric forms of PKM2, which is sensitive to the availability of fructose-1,6-bisphosphate plays an important role in cancer metabolism and is reportedly altered by various mechanisms. PKM2 contains a phosphorylated tyrosine peptide-binding domain that is absent in the M1 splice form.33 Binding of phosphorylated tyrosine peptide to PKM2 leads to the release of fructose-1,6-bisphosphate resulting in a shift in the equilibrium towards the less active dimeric form. The mRNA for PKM1 and PKM2 are produced from same pre-mRNA by alternative splicing with the inclusion of exon9 for PKM1 and exon10 for PKM2. Exon10 represents a phosphorylated tyrosine peptide binding site and it has been reported that heterogeneous nuclear ribonucleo-protein (hnRNP) proteins, poly-pyrimidine tract binding proteins (PTB, also known as hnRNPI), hnRNPA1 and hnRNPA2 bind to sequences flanking exon9, resulting in exon10 inclusion leading to the expression of M2 splice isoform of pyruvate kinase. Transcription of these three factors is up-regulated by oncogenic transcription factor c-Myc, which has been reported to be dysregulated in various cancers.33 In another regulatory mechanism, phosphorylation of PKM2 at tyrosine-105 by fibroblast growth factor receptor type-1 leads to the inhibition of the formation of PKM2 tetramer through disruption of its interaction with fructose-1,6-bisphosphate.34 Phosphorylation of tyrosine-105 is common in several cancers and has been reported to be important for cancer growth. Generation of reactive oxygen species (ROS) can also affect the activity of PKM2 as it has been found that oxidation of cysteine-358 by ROS reduces PKM2 activity.35 These regulatory mechanisms are exploited to various extents by cancer cells to change the levels of glycolytic intermediates in such a way to support proliferative state maintained by cancer cells. While PKM2 seems to be an ideal target for targeted therapy, inhibition of PKM2 leads to better proliferation in many cancers. This is because PKM2 inhibition leads to glycolytic intermediate accumulation, which leads to higher flux into biosynthetic pathways. In this case, targeting the allosteric mechanisms, which regulate the PKM2 activity, might prove to be beneficial. Thus understanding the role of metabolic changes in supporting cancer development is very important in developing strategies based on inhibition of cancer metabolism.2.5. Pyruvate dehydrogenase (PDH)
Metabolic fate of pyruvate affects many biochemical properties of cells like substrate preference, red-ox state, etc. Entry of pyruvate into mitochondria is mediated by PDH-complex. Cancer cells prevent the entry of pyruvate into mitochondria and shuttle the flux into lactic acid production. Pyruvate dehydrogenase activity is regulated by pyruvate dehydrogenase kinase (PDK) that inactivates PDH through phosphorylation and pyruvate dehydrogenase phosphatase (PDP) that activates PDH through dephosphorylation.36 Four isoforms of PDK have been identified (PDK1-4) and PDK3 is the most catalytically active and less sensitive to inhibition by high pyruvate levels. Hypoxia leads to higher PDK3 expression thus promoting aerobic glycolysis by suppression of oxidative phosphorylation in oxygen deprived conditions. PDK1 is established to be one of the direct targets of hypoxia inducible factor (HIF1).37 Stabilization of HIF1 by hypoxia increases the expression of PDK1. Dysregulated c-Myc and HIF synergistically induce expression of PDK1 in cancer cells.38 Inhibition of PDK activity by dichloroacetate (DCA) leads to increased PDH activity and increased oxidative phosphorylation.39 Although use of DCA as independent chemotherapeutic agent is limited combinational administration of DCA with other anti-cancer agents like omeprazole, tamoxifen and 5-flurouracil has provided promising results.402.6. Lactate dehydrogenase (LDH)
The fate of pyruvate depends on the availability of oxygen in normal cells. When mitochondrial function is diminished, regeneration of NAD+ for glycolytic requirements is mediated by conversion of pyruvate to lactate catalysed by LDH. There are several isoforms of LDH. Over-expression of LDH5 has been reported in several cancer types.41,42 Hypoxic conditions in solid tumours and acidic pH in tumour microenvironment are related to high expression of LDH5.42 LDH is up-regulated by accumulation of HIF1α and HIF2α. Up-regulation of LDH by HIF is understandable because under hypoxic conditions, the regeneration of NAD+ for glycolysis is solely dependent on LDH enzymes. LDHA is over-expressed in certain gliomas and inhibition of LDHA has been found to retard cancer progression.43 Fork-head box protein (FOXM1) is reported to up-regulate the expression of LDHA in pancreatic cancer at the transcription level by directly binding to LDHA gene promoter.44 Krüppel-like factor 4 [KLF4], a tumour suppressor which is dysregulated in pancreatic cancers and several other cancers, has been reported to down-regulate the expression of LDHA.45 LDHC, a germ cell specific enzyme has been identified in human cancer cells.46 Lactate is the preferred substrate for LDHC and thus expression of LDHC may serve as a metabolic rescue mechanism to generate ATP from lactate. However, the mechanism by which LDHC escapes from transcriptional repression is yet to be understood. Reduced LDH activity retards the ability of cancer cells to regenerate NAD+ necessary for glycolysis thus limiting the ATP generation by glycolysis. Knock-down of LDH leads to increase in oxidative phosphorylation, increase in oxidative stress and reduced proliferation thus making it a promising therapeutic target in treating cancer. Inhibition of lactate formation in cancers leads to reduced lactate accumulation in tumour microenvironment. Lactate accumulation has been linked to effects like reverse Warburg effect, vascular endothelial lactate shuttle and metabolic symbiosis.47 Extracellular acidosis caused by accumulation of lactate under hypoxic conditions gives rise to physiological barriers thus contributing to cancer drug resistance.47 Targeting LDH may therefore, have multiple advantages and can help in increasing the efficacy of other anti-cancer agents.2.7. Isocitrate dehydrogenase (IDH)
Isocitrate dehydrogenase (IDH) catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate with NADP+ or NAD+ as electron acceptor. NAD+ dependent IDH3 catalyzes the irreversible decarboxylation of isocitrate to α-ketoglutarate while NADP+ dependent IDH1 and IDH2 catalyze the reductive carboxylation of α-ketoglutarate to isocitrate. IDH3 is a multi-subunit enzyme localized on inner mitochondrial membrane and plays a major role in aerobic cellular energetics. IDH1 is localized on cytosol and peroxisomes and IDH2 is localized in the inner mitochondrial membrane.48 Several functional mutations in the active site of IDH1 and IDH2 enzymes have been reported in several cancer types like glioblastoma.48,49 IDH1-Arg 132 and IDH2-Arg140 mutations have been reported in acute myeloid leukemia (AML).50,51 These mutations lead to change in the function of IDH1 and IDH2 where the mutated enzymes convert α-ketoglutarate to 2-hydroxyglutarate (2-HG).52 Mutations in IDH1 and IDH2 have been linked to HIF-mediated cancer promotion in some recent studies.53 A possible link between IDH mutation and cancer development may be due to depletion of NADPH and α-ketoglutarate. α-Ketoglutarate is an essential substrate for prolyl hydroxylase (PHD) enzymes that regulate the degradation of HIF1α. Depletion of NADPH may also cause oxidative stress leading to other mutations. Normal cells produce very low amounts of 2-HG while cancer cells produce abnormal quantities of 2-HG. This property of IDH-dependent cancer can be used for diagnostic applications. 2-HG has been reported to inhibit a number of enzymes of 2-oxoglutarate-dioxygenase class.54 These enzymes are involved in a variety of cellular processes including epigenetic modification. However, a complete understanding of the role played by 2-HG in cancer development is required in order to come up with novel therapeutic strategies targeting IDH.2.8. Succinate dehydrogenase and fumarate hydratase
Although mitochondrial dysfunction has been linked to cancer development since the discovery of Warburg effect, direct biochemical evidences for the relationship between mitochondrial dysfunction and development of cancer have started to accumulate in recent years. Succinate dehydrogenase (SDH) and fumarate hydratase (FH) are enzymes that catalyze two successive reactions in TCA cycle (succinate to fumarate and fumarate to malate respectively). Dysfunction of these two enzymes has been linked to development of various cancers in recent findings.55 Germline mutations in FH gene has been linked to skin, renal and uterus cancers.56–58 FH inhibition by 3-nitro propionic acid causes stabilization and up-regulation of HIF.59 Germline inactivating mutations in SDH gene has been linked to paraganglioma.60 Findings that link SDH and FH dysfunction to cancer development provides evidence that metabolites can be oncogenic. HIF has been reported to play a crucial role in development and survival of cancers with SDH and FH dysfunction.59 Loss of SDH or FH function leads to accumulation of respective substrates succinate and fumarate. Both metabolites are capable of passing through inner mitochondrial membrane and entering cytosol where they inhibit the activity of PHD enzymes.59,61 These enzymes are responsible for the degradation of HIF-1α under normoxia condition. Succinate is not only a substrate for SDH but also the product of reaction catalyzed by PHD enzymes. Fumarate due to its structural similarities with succinate also acts as an inhibitor of PHD enzyme. The role of HIF in cancer development and survival has been well documented in literature and reviewed elsewhere.62 Loss of SDH or FH function leads to a pseudo hypoxia even in the presence of oxygen thus linking mitochondrial dysfunction to cancer development. Although both SDH and FH loss seems to be functionally similar, the overall phenotype of cancers differ. This might be because the accumulated succinate and fumarate inhibit different PHD enzymes with different specificity.3. Branching pathways
Changes in energy metabolism are complimented by changes in branching pathways thus leading to higher levels of biosynthesis in cancer cells. In the following section, few examples of complementation between changes in energy metabolism and biosynthesis in cancer are discussed.3.1. Cancer nucleotide metabolism
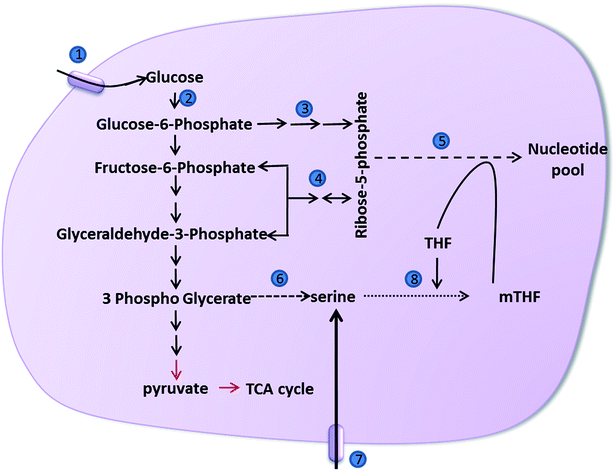
Nucleotide synthesis in cancer cells is almost exclusively dependent de novo pathway rather than the salvage pathway due to their proliferative nature. Synthesis of nucleotides by de novo pathway requires intermediates from glycolysis as precursors. Ribose-5-phosphate (R5P) is obtained from pentose-phosphate pathway (PPP), which utilizes glycolytic intermediates for synthesis of R5P. The reversible nature of non-oxidative pentose-phosphate pathway renders it highly sensitive to changes in the levels of glycolytic intermediates. Many cancer cells possess the pyruvate kinase M2 splice isoform leading to accumulation of glycolytic intermediates. Therefore, they utilize the non-oxidative branch extensively for R5P synthesis. Under hypoxic conditions PFK activity is reduced by modification with O-linked β-N-acetyl glucosamine in a serine residue.63 This modification is a fail-safe used by cancer cells to generate NADPH through PPP. Cancers depend on PPP for higher nucleotide synthesis and NADPH generation of reductive biosynthesis. Thus strategies to decrease the flux through PPP can be used in combination with nucleotide analogues that are used in conventional chemotherapy to overcome resistance. Fig. 2 depicts a schematic representation of various pathways involved in cancer nucleotide metabolism.3.2. One carbon metabolism
One carbon metabolism centred on folate has an important role in utilizing de novo or exogenous input of amino acids, glucose and vitamins to generate a wide range of output such as lipids, nucleotides, redox status maintenance apart from substrates for methylation reactions. The folate cycle and the methionine cycle together contribute to the one carbon-metabolism and extensive crosstalk exists between these cycles. Carbon units for one-carbon metabolism are provided serine derived from a branch of glycolysis and serine imported by facilitated transport.Cancer cells use approximately 10% of the glycolytic intermediate 3-phosphoglycerateto produce serine precursors.64 Accumulation of glycolytic intermediates leads to increased serine production. They use phosphoglycerate-dehydrogenase (PHGDH) to oxidize 3-phosphoglycerate into 3-phosphohydroxypyruvate. This is followed by a transamination reaction mediated by phosphoserine-aminotransferase (PSAT1) and phosphate ester hydrolysis (PSPH) reactions to yield serine. Serine is converted to glycine by transfer of its side chain to folate, catalysed by hydroxymethyl-transferase (SHMT). PHGDH is normally found to be up-regulated in several cancers like melanoma and triple-negative breast cancer.65,66 PHGDH suppression results in inhibition of cell proliferation, even in the presence of exogenous serine, suggesting that PHGDH, besides controlling intracellular serine levels participates in other metabolic processes.66 Recent studies show that cancer cells selectively consume exogenous serine in the absence of which, exogenous glycine uptake was not able to support nucleotide synthesis.67 Higher concentrations of glycine were found to exert a negative effect on cell proliferation due to the conversion of glycine to serine, diminishing the one-carbon pool.67 It has also been found that a glycine cleavage system, which produces CO2, NH3 and a carbon unit used for the methylation of THF through a glycine dehydrogenase-mediated cleavage of glycine, becomes active in certain cells, thus charging the folate cycle.68 Aldol cleavage reaction of threonine catalysed by threonine dehydrogenase (TDH) and glycine C-acetyl-transferase (GCAT) to form glycine.69 Threonine thus enters the folate cycle via glycine cleavage. Abundant intracellular serine allosterically activates PKM2, which helps to utilize glucose through aerobic glycolysis. Serine deprivation reduces PKM2 activity by diverting carbon units from the pyruvate to serine biosynthetic pathways70,71 In addition, the members of the p53 family namely, p53, p63 and p73 have been shown to activate the expression of glutaminase-2 (GLS-2), which in turn promotes glutaminolysis, thereby interfering with serine biosynthesis.72–76 During serine starvation, p53–p21 axis is activated leading to cell cycle arrest, promoting cell survival by efficient channelling of depleted serine stores to glutathione synthesis.77 Recent studies show that TAp73, a member of p53 family activates serine biosynthesis and increases intracellular levels of serine, glycine, and GSH.78 Managing serine levels in cells can therefore be an attractive anti-cancer strategy.
Methylation of dUMP (deoxyuridine monophosphate) by thymidylate synthase to produce dTMP (deoxythymidine monophosphate) requires methylated-tetrahydrofolate (mTHF) as a methyl donor. Purine biosynthesis also requires a folate pool. Methionine adenylation produces S-adenosyl methionine a methyl donor for methylation of histone, DNA and RNA, lysine, arginine, and polyamine synthesis. Current drugs that target folate pathway are mostly thymidylate synthase inhibitors. Not much research in the direction of exploiting the role of glycine and serine in one carbon metabolism has been carried out.
3.3. Glutaminolysis
Glutamine contributes to energy metabolism through the citric acid (TCA) cycle and also provides nitrogen and carbon skeletons for cancer cells that are actively proliferating. The extent to which the cancer cells utilize glucose and glutamine depends on its genotype. It is now established that transformation of pyruvate to acetyl-CoA catalysed by pyruvate dehydrogenase and its subsequent entry into mitochondria is down-regulated in many cancers. But, cancer cells use a truncated TCA cycle fuelled by glutamine to generate biosynthetic precursors. α-Ketoglutarate produced in mitochondrial glutamine metabolism is used to produce citrate, which is then transported to cytosol where it is converted to acetyl CoA by ATP-citrate lyase thus providing acetyl CoA for de novo fatty acid synthesis.79Glutamine can be synthesized from glutamate and ammonia by glutamine synthetase. Cancer cells with elevated glutamine synthetase therefore, do not require exogenous glutamine. Cell internalization of L-glutamine and its quick efflux in the presence of essential amino acids (EAA) occurs via the bidirectional transporter SLC7A5/SLC3A2.80 Transport of glutamine across cell membrane is recognized as the rate-limiting step in glutamine metabolism and found to regulate mTOR activation. SLC1A5 regulates glutamine uptake, loss of which has been found to inhibit cell growth and activate autophagy.80 Cells when cultured for more than two days in glutamine-containing medium exhibited increase in autophagy, which was neither due to nutrient depletion nor inhibition of mTOR. Conditioned medium obtained from these cells was found to contain a volatile factor, triggering autophagy in secondary cultures. This factor was identified to be ammonia derived from glutamine by glutaminolysis, which also protected cells from tumour necrosis factor alpha induced cell death.81 Interestingly, ammonia has been shown to be sufficient for long-term survival and proliferation of human Hepatoma Cell (Hep3B) in the absence of glutamine.82 It was also shown that glutamine independent derivative of Hep3B exhibits high levels of glutamine synthetase. These evidences suggest that glutamate supplies the nitrogen rather than the carbon skeleton for cell proliferation. However, similar experiments on other cells lines demonstrated glutamine dependency suggesting that the mode of glutamine or utilization of glucose is influenced by the metabolic state of the cancer cells.82
Myc oncogene is a well-known inducer of both aerobic glycolysis and glutaminolysis. It was observed that more potent triggering of cell death is induced by glutamine withdrawal rather than glucose withdrawal in Myc-transformed cells.83 Myc directly regulates genes involved in glutamine metabolism and induces expression of mitochondrial glutaminase, an enzyme that converts glutamine to glutamate.84,85 Early stage mammalian embryos are shown to utilize aerobic glycolysis and glutaminolysis.86 This suggests that highly undifferentiated cancer cells can revert to aerobic glycolysis and glutaminolysis. The role of glutamine varies depending upon genetic and epigenetic composition of various cancer cells. In some cases of cancer and mammalian cell types, isotopic studies demonstrate a role for glutamine in providing anaplerotic carbons.87,88 Reducing the activity of glutaminase using bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl) ethyl sulphide has been shown to cause an increased accumulation of glycolytic intermediates for survival in IDH1-mutated glioblastoma cells.89 Thus simultaneous inhibition of glutaminolysis and glycolysis might prove to be an efficient strategy against cancers with IDH1 mutation. Fig. 3 shows a cartoon on the steps involved in the glutaminolysis pathway.
4. Significance of metabolic changes
In a multi-cellular organism, the freedom of each cell to replicate independently is compromised and they grow in a controlled and co-ordinate manner regulated by signalling mechanisms. The metabolic requirements of proliferative and quiescent cells are distinctly different. Cellular energy metabolism is a key process not only because it provides energy required to maintain the integrity of the cells but also provides precursors for the biosynthesis of building blocks required for producing macromolecular structures (nucleic acids, proteins, etc.). It is well known fact that cancer cells become self-sufficient with respect to signals required for growth i.e. they are transformed into an independent entity whose growth is not controlled by signalling mechanisms as observed in normal cells.3 Once cells overcome the barriers of regulated growth, Darwinian selection emerges as a key player in their transformation into cancerous cells. A proliferating cell needs to meet various requirements to double its cell mass. Transformed cells that do not satisfy these requirements perish, and cells best suited for rapid proliferation are selected. This natural selection may occur during the dormant stage of the cancer cells suggested by Warburg during which cells with higher glycolytic capacity are selected. But the reason for selection of the energy inefficient lactic acid fermentation over oxidative phosphorylation leaves one baffled. The answers to this intriguing choice may lie with the cellular metabolic requirements – few facets of which are discussed in the following sections.4.1. Finding the balance between energy generation and biosynthesis
An important requirement for a cancer cell to retain its proliferative state is to maintain a balance between the energy generation and biosynthesis. In quiescent cells from which cancer cells arise there is a huge imbalance between these two processes. For example, the net reaction of fatty acid (palmitate) production by reductive biosynthesis is as follows:| 8Acetyl-CoA + 14NADPH + 7ATP → Palmitate + 14NADP+ + 8CoA + 6H2O + 7ADP + 7Pi |
One glucose molecule can yield 32 ATP molecules or 2 NADPH. It can be inferred that one glucose molecule can generate enough ATP to produce about five palmitate molecules while seven glucose molecules are required to generate sufficient NADPH for producing a single palmitate molecule. Thus, there is nearly a 35-fold imbalance between the generation of ATP and NADPH required for fatty acid synthesis. Decreasing the flux into TCA cycle and increasing the flux into pentose phosphate pathway may overcome this imbalance. This seems to be in operation in many cancer cells. In cancer cells, glucose carbon is shuttled into various biosynthetic pathways unlike the normal cells where most of the glucose carbon enters the TCA cycle to be eliminated as CO2. Also cancer cells use a truncated TCA cycle fuelled by glutamine to produce citrate that is exported to the cytosol where it forms oxaloacetate and acetyl-CoA in a reaction catalysed by ATP-dependent citrate lyase. This acetyl-CoA is further utilized for fatty acid synthesis by cancer cells.
4.2. Early selection of cells with higher biosynthetic ability
During the early stages of cancer development, it has been reported that several signalling pathways, which regulate cell cycle are dysregulated thereby forcing the cell to remain in a proliferative state.90 Such proliferation without sufficient nucleotide pool leads to chromosomal instability caused due to stress on DNA replication. In cervical cancers caused by human papilloma virus (HPV) Rb-E2F pathway is dysregulated. Aberrant activation of Rb-E2F leads to DNA double strand breaks, which in turn leads to senescence and apoptosis. This is due to imbalance between the cell cycle and nucleotide biosynthesis. It has been shown that this problem of genomic instability can be overcome by exogenous supply of nucleotides but this reduced the oncogene-induced transformation.90 Lack of selective demand for higher rate of nucleotide biosynthesis might be the reason for the drastic reduction in the oncogene-induced transformation. It has also been shown that up-regulation of nucleotide biosynthesis by over-expression of c-Myc also helps to rescue the cells from chromosomal instability.90 Though the importance of various cellular mechanisms that are altered during the cancer development is not understood completely, it is evident that certain change such as establishment of glycolytic phenotype, which helps to increase nucleotide biosynthesis, are essential.4.3. Using lipid synthesis to maintain pH and redox state of cells
Embryonic cells rely on de novo fatty acid synthesis while it is suppressed in adult cells due to the surplus availability of nutritional fatty acids.91 Interestingly, cancer cells also tend to rely on de novo fatty acid synthesis. While higher fatty acid requirement in the cancerous cells is understandable, it is puzzling to note that this need is not satisfied by the nutritional fatty acids present in the blood stream. As molecular mechanisms that establish the glycolytic phenotype and their involvement in fatty acid synthesis are now understood to a reasonable extent, it is likely that de novo fatty acid synthesis may have some interesting roles to play in cancer metabolism. Increased glucose uptake leads to increased lactate production resulting in a rise in the intracellular pH. Lipid metabolism can be used as a carbon sink to avoid excess lactate production thus helping to maintain intracellular pH. In another perspective, NADP+ produced during fatty acid synthesis can be used to maintain the redox state of cells. Some hypoxia tolerant organisms use NADP+ as electron acceptor in the absence of oxygen and cancer cells may adopt the same strategy under hypoxic conditions.92 It has been proposed that under hypoxic conditions, NADP+ can contribute to availability of cytosolic NAD+.93 The enzyme IDH1 uses the NADP+ in generation of α-ketoglutarate, which is then transported into the mitochondria where the reverse reaction is catalysed by IDH-2 with generation of NADP+. This NADP+ is used as electron acceptor in the absence of oxygen to maintain the NADH/NAD+ ratio by nucleotide trans-hydrogenase.93 This NAD+ can be made available for glycolysis.4.4. Mitochondrial binding of hexokinase and evasion from apoptosis
More than half of the HK expressed in cancer cells is associated with mitochondria thereby directly coupling glycolysis to oxidative phosphorylation. This helps HK to become insensitive to feedback inhibition by G6P and provides a preferential access for one of its substrates ATP (generated by oxidative phosphorylation). It has been reported that HK binding to VDAC protects cancer cell against apoptosis mediated by interaction of apoptotic proteins with VDAC.94,95 Outer mitochondrial membrane permeability, which is controlled by VDAC, plays a crucial role in apoptosis. VDAC has been reported to be important in release of cytochrome-c from mitochondria during apoptosis.96 It has also been reported that it interacts with pro- and anti-apoptotic proteins of Bcl-2 family.97 HK2 binding to mitochondria inhibits Bax-induced cytochrome-c release.95 The mechanism by which binding of HK to VDAC promotes cell survival is not completely understood but a plausible explanation might be that the binding of HK to VDAC may inhibit its interactions with apoptotic signals thereby preventing the release of the pro-apoptotic cytochrome-c.5. Advantage of targeting cancer metabolism
Sufficient body of literature exists to suggest that metabolic changes conferring multiple advantages to the cancer cells are selected during cancer progression. Although the origin of cancer is different in various forms of cancer, changes in energy metabolism are similar in most cancers and these alterations play a central role in making cancer cells self-sufficient. Targeting energy metabolism as a strategy to treat cancer can be advantageous, in that it not only affects the cellular energy status but also affect biosynthetic ability of cells to support the proliferation of cancer cells. Proliferative property of cancer cells is exploited by most of the current cancer treatment strategies. They target DNA replication and aim to induce DNA damage but these strategies are limited due to development of drug resistance. One of the major causes for the onset of drug resistance in cancer cells is due to the selection of cells with higher biosynthetic capability from a heterogeneous population of cancer cells. On the other hand, targeting energy metabolism, specifically glycolysis, leads to selection of cells, which proliferate at a slower rate and have a functional OXPHOS pathway. Complementing drugs that target the proliferative property with glycolytic inhibitors have been proved to be very efficient. One best example of this concept is the use of the HK inhibitor, 2-deoxy glucose (2-DG), to increase the efficacy of the chemotherapeutic agent, 5-flurouracil (5-FU).98 2DG is a glucose analogue, which gets phosphorylated by HK but does not participate in subsequent reactions of glycolysis thus leading to ATP depletion. One of the mechanisms by which 2-DG potentiates the activity of 5-FU is that ATP depletion leads to activation of AMPK pathway and lowers the levels of phosphorylated Akt which in turn leads to leads to lower biosynthetic ability of treated cells.Another perspective where cancers are viewed as metabolic steady states established by genetic changes provides some interesting strategies in treating cancers. One such strategy based on planned diet is to replace glucose and glutamine, which are favoured nutrients for cancer cells, with alternatives like ketone bodies. Ketone bodies are generally produced in livers from lipids during the time of carbohydrate limitation to be used as energy source by the body. Cells convert these compounds into Acetyl CoA, which is fed through TCA cycle for generation of energy.99 As cancer cells depend heavily on glycolysis for generation of ATP, it has been reported that ketone body-rich diets lead to reduced glycolytic flux and reduced cancer cell proliferation.100,101
6. Conclusion
The energy requirements of quiescent and proliferating cells are quite different. While TCA cycle and oxidative phosphorylation are used by non-proliferative cells for harvesting energy from glucose efficiently, proliferating cells resort to anaerobic glycolysis for various purposes like maintaining high rates of nucleotides synthesis, escaping apoptosis etc., and use TCA cycle for generation of biosynthetic precursors by fuelling TCA anaplerotically. The establishment and maintenance of proliferative metabolism requires number genetic changes. These changes need to be sequential because an imbalance between biosynthesis and cell division can be seriously damaging to proliferating cells. Although the sequential order in which these changes should be established is relatively unknown, there seems to be a convergent evolution when it comes to changes in cancer energy metabolism. This is because the metabolic requirements of all proliferation cells are similar. This may be the reason why embryonic cells have metabolic state similar to that of cancer cells. Cancers are not caused just by mutations but they evolve under a selective pressure to maintain higher rate of proliferation and to survive harsh environments. Treating cancer requires understanding of cancer as an altered metabolic phenotype. Identification of novel targets in the cancer metabolic pathways represents an emerging paradigm in the field of cancer therapeutics.Acknowledgements
The authors acknowledge FIST, Department of Science & Technology, India (SR/FST/LSI-327/2007 & SR/FST/LSI-058/2010) for financial support. SASTRA University is also acknowledged for infrastructure support.References
- O. Warburg, Science, 1956, 123, 309–314 CAS.
- H. G. Crabtree, Biochem. J., 1928, 22, 1289–1298 CAS.
- D. Hanahan and R. A. Weinberg, Cell, 2000, 100, 57–70 CrossRef CAS.
- M. Hatanaka, Biochim. Biophys. Acta, 1974, 355, 77–104 CAS.
- C. Chen, N. Pore, A. Behrooz, F. Ismail-Beigi and A. Maity, J. Biol. Chem., 2001, 276, 9519–9525 CrossRef CAS PubMed.
- A. L. Edinger, Biochem. J., 2007, 406, 1–12 CrossRef CAS PubMed.
- T. G. Sommermann, K. O'Neill, D. R. Plas and E. Cahir-McFarland, Cancer Res., 2011, 71, 7291–7300 CrossRef CAS PubMed.
- B. B. Kahn, J. Clin. Invest., 1992, 89, 1367–1374 CrossRef CAS PubMed.
- A. L. Olson and J. E. Pessin, Annu. Rev. Nutr., 1996, 16, 235–256 CrossRef CAS PubMed.
- K. K. Chan, J. Y. Chan, K. K. Chung and K. P. Fung, J. Cell. Biochem., 2004, 93, 1134–1142 CrossRef CAS PubMed.
- L. Sui, Y. Dong, Y. Watanabe, F. Yamaguchi, N. Hatano, I. Tsukamoto, K. Izumori and M. Tokuda, Int. J. Oncol., 2005, 27, 907–912 CAS.
- E. Bustamante, H. P. Morris and P. L. Pedersen, J. Biol. Chem., 1981, 256, 8699–8704 CAS.
- E. Bustamante and P. L. Pedersen, Proc. Natl. Acad. Sci. U. S. A., 1977, 74, 3735–3739 CrossRef CAS.
- S. P. Mathupala, A. Rempel and P. L. Pedersen, J. Bioenerg. Biomembr., 1997, 29, 339–343 CrossRef CAS.
- J. P. Tuttle and J. E. Wilson, Biochim. Biophys. Acta, 1970, 212, 185–188 CrossRef CAS.
- R. A. Nakashima, J. Bioenerg. Biomembr., 1989, 21, 461–470 CrossRef CAS.
- M. Rho, J. Kim, C. D. Jee, Y. M. Lee, H. E. Lee, M. A. Kim, H. S. Lee and W. H. Kim, Anticancer Res., 2007, 27, 251–258 CAS.
- P. L. Pedersen, S. Mathupala, A. Rempel, J. F. Geschwind and Y. H. Ko, Biochim. Biophys. Acta, 2002, 1555, 14–20 CrossRef CAS.
- L. Sun, S. Shukair, T. J. Naik, F. Moazed and H. Ardehali, Mol. Cell. Biol., 2008, 28, 1007–1017 CrossRef CAS PubMed.
- G. N. Bijur and R. S. Jope, J. Neurochem., 2003, 87, 1427–1435 CrossRef CAS.
- S. J. Pilkis and T. H. Claus, Annu. Rev. Nutr., 1991, 11, 465–515 CrossRef CAS PubMed.
- E. Van Schaftingen, L. Hue and H. G. Hers, Biochem. J., 1980, 192, 897–901 CAS.
- S. Ros and A. Schulze, Canc. Metabol., 2013, 1, 8 CrossRef PubMed.
- O. Minchenko, I. Opentanova, D. Minchenko, T. Ogura and H. Esumi, FEBS Lett., 2004, 576, 14–20 CrossRef CAS PubMed.
- T. Atsumi, J. Chesney, C. Metz, L. Leng, S. Donnelly, Z. Makita, R. Mitchell and R. Bucala, Cancer Res., 2002, 62, 5881–5887 CAS.
- J. Chesney, R. Mitchell, F. Benigni, M. Bacher, L. Spiegel, Y. Al-Abed, J. H. Han, C. Metz and R. Bucala, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 3047–3052 CrossRef CAS.
- R. C. Osthus, H. Shim, S. Kim, Q. Li, R. Reddy, M. Mukherjee, Y. Xu, D. Wonsey, L. A. Lee and C. V. Dang, J. Biol. Chem., 2000, 275, 21797–21800 CrossRef CAS PubMed.
- G. E. Staal, A. Kalff, E. C. Heesbeen, C. W. van Veelen and G. Rijksen, Cancer Res., 1987, 47, 5047–5051 CAS.
- M. Lopez-Lazaro, Anti Canc. Agents Med. Chem., 2008, 8, 305–312 CrossRef CAS.
- H. R. Christofk, M. G. Vander Heiden, M. H. Harris, A. Ramanathan, R. E. Gerszten, R. Wei, M. D. Fleming, S. L. Schreiber and L. C. Cantley, Nature, 2008, 452, 230–233 CrossRef CAS PubMed.
- K. Ashizawa, M. C. Willingham, C. M. Liang and S. Y. Cheng, J. Biol. Chem., 1991, 266, 16842–16846 CAS.
- S. Mazurek, C. B. Boschek, F. Hugo and E. Eigenbrodt, Semin. Cancer Biol., 2005, 15, 300–308 CrossRef CAS PubMed.
- C. J. David, M. Chen, M. Assanah, P. Canoll and J. L. Manley, Nature, 2010, 463, 364–368 CrossRef CAS PubMed.
- T. Hitosugi, S. Kang, M. G. Vander Heiden, T. W. Chung, S. Elf, K. Lythgoe, S. Dong, S. Lonial, X. Wang, G. Z. Chen, J. Xie, T. L. Gu, R. D. Polakiewicz, J. L. Roesel, T. J. Boggon, F. R. Khuri, D. G. Gilliland, L. C. Cantley, J. Kaufman and J. Chen, Sci. Signaling, 2009, 2, ra73 CrossRef PubMed.
- D. Anastasiou, G. Poulogiannis, J. M. Asara, M. B. Boxer, J. K. Jiang, M. Shen, G. Bellinger, A. T. Sasaki, J. W. Locasale, D. S. Auld, C. J. Thomas, M. G. Vander Heiden and L. C. Cantley, Science, 2011, 334, 1278–1283 CrossRef CAS PubMed.
- M. C. Sugden and M. J. Holness, Arch. Physiol. Biochem., 2006, 112, 139–149 CrossRef CAS PubMed.
- J. W. Kim, I. Tchernyshyov, G. L. Semenza and C. V. Dang, Cell Metab., 2006, 3, 177–185 CrossRef PubMed.
- J. W. Kim, P. Gao, Y. C. Liu, G. L. Semenza and C. V. Dang, Mol. Cell. Biol., 2007, 27, 7381–7393 CrossRef CAS PubMed.
- E. D. Michelakis, L. Webster and J. R. Mackey, Br. J. Cancer, 2008, 99, 989–994 CrossRef CAS PubMed.
- T. Ishiguro, R. Ishiguro, M. Ishiguro and S. Iwai, Hepato-Gastroenterology, 2012, 59, 994–996 CAS.
- M. I. Koukourakis, A. Giatromanolaki, E. Sivridis, G. Bougioukas, V. Didilis, K. C. Gatter and A. L. Harris, Br. J. Cancer, 2003, 89, 877–885 CrossRef CAS PubMed.
- M. I. Koukourakis, A. Giatromanolaki, C. Simopoulos, A. Polychronidis and E. Sivridis, Clin. Exp. Metastasis, 2005, 22, 25–30 CrossRef CAS PubMed.
- A. Le, C. R. Cooper, A. M. Gouw, R. Dinavahi, A. Maitra, L. M. Deck, R. E. Royer, D. L. Vander Jagt, G. L. Semenza and C. V. Dang, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 2037–2042 CrossRef CAS PubMed.
- J. Cui, M. Shi, D. Xie, D. Wei, Z. Jia, S. Zheng, Y. Gao, S. Huang and K. Xie, Clin. Cancer Res., 2014, 20, 2595–2606 CrossRef CAS PubMed.
- M. Shi, J. Cui, J. Du, D. Wei, Z. Jia, J. Zhang, Z. Zhu, Y. Gao and K. Xie, Clin. Cancer Res., 2014, 20, 4370–4380 CrossRef CAS PubMed.
- M. Koslowski, O. Tureci, C. Bell, P. Krause, H. A. Lehr, J. Brunner, G. Seitz, F. O. Nestle, C. Huber and U. Sahin, Cancer Res., 2002, 62, 6750–6755 CAS.
- J. R. Doherty and J. L. Cleveland, J. Clin. Invest., 2013, 123, 3685–3692 CrossRef CAS PubMed.
- D. Krell, M. Assoku, M. Galloway, P. Mulholland, I. Tomlinson and C. Bardella, PLoS One, 2011, 6, e19868 CAS.
- K. Ichimura, D. M. Pearson, S. Kocialkowski, L. M. Backlund, R. Chan, D. T. Jones and V. P. Collins, Neuro-Oncology, 2009, 11, 341–347 CrossRef CAS PubMed.
- J. R. Prensner and A. M. Chinnaiyan, Nat. Med., 2011, 17, 291–293 CrossRef CAS PubMed.
- L. Dang, S. Jin and S. M. Su, Trends Mol. Med., 2010, 16, 387–397 CrossRef CAS PubMed.
- D. Ye, S. Ma, Y. Xiong and K. L. Guan, Cancer Cell, 2013, 23, 274–276 CrossRef CAS PubMed.
- P. S. Ward, J. Patel, D. R. Wise, O. Abdel-Wahab, B. D. Bennett, H. A. Coller, J. R. Cross, V. R. Fantin, C. V. Hedvat, A. E. Perl, J. D. Rabinowitz, M. Carroll, S. M. Su, K. A. Sharp, R. L. Levine and C. B. Thompson, Cancer Cell, 2010, 17, 225–234 CrossRef CAS PubMed.
- P. Koivunen, M. Hirsila, A. M. Remes, I. E. Hassinen, K. I. Kivirikko and J. Myllyharju, J. Biol. Chem., 2007, 282, 4524–4532 CrossRef CAS PubMed.
- P. J. Pollard, J. J. Briere, N. A. Alam, J. Barwell, E. Barclay, N. C. Wortham, T. Hunt, M. Mitchell, S. Olpin, S. J. Moat, I. P. Hargreaves, S. J. Heales, Y. L. Chung, J. R. Griffiths, A. Dalgleish, J. A. McGrath, M. J. Gleeson, S. V. Hodgson, R. Poulsom, P. Rustin and I. P. Tomlinson, Hum. Mol. Genet., 2005, 14, 2231–2239 CrossRef CAS PubMed.
- N. A. Alam, S. Olpin and I. M. Leigh, Br. J. Dermatol., 2005, 153, 11–17 CrossRef CAS PubMed.
- L. Stewart, G. M. Glenn, P. Stratton, A. M. Goldstein, M. J. Merino, M. A. Tucker, W. M. Linehan and J. R. Toro, Arch. Dermatol., 2008, 144, 1584–1592 Search PubMed.
- J. R. Toro, M. L. Nickerson, M. H. Wei, M. B. Warren, G. M. Glenn, M. L. Turner, L. Stewart, P. Duray, O. Tourre, N. Sharma, P. Choyke, P. Stratton, M. Merino, M. M. Walther, W. M. Linehan, L. S. Schmidt and B. Zbar, Am. J. Hum. Genet., 2003, 73, 95–106 CrossRef CAS PubMed.
- J. S. Isaacs, Y. J. Jung, D. R. Mole, S. Lee, C. Torres-Cabala, Y. L. Chung, M. Merino, J. Trepel, B. Zbar, J. Toro, P. J. Ratcliffe, W. M. Linehan and L. Neckers, Cancer Cell, 2005, 8, 143–153 CrossRef CAS PubMed.
- B. Pasini and C. A. Stratakis, J. Intern. Med., 2009, 266, 19–42 CrossRef CAS PubMed.
- M. A. Selak, S. M. Armour, E. D. MacKenzie, H. Boulahbel, D. G. Watson, K. D. Mansfield, Y. Pan, M. C. Simon, C. B. Thompson and E. Gottlieb, Cancer Cell, 2005, 7, 77–85 CrossRef CAS PubMed.
- G. L. Semenza, Nat. Rev. Cancer, 2003, 3, 721–732 CrossRef CAS PubMed.
- W. Yi, P. M. Clark, D. E. Mason, M. C. Keenan, C. Hill, W. A. Goddard 3rd, E. C. Peters, E. M. Driggers and L. C. Hsieh-Wilson, Science, 2012, 337, 975–980 CrossRef CAS PubMed.
- R. J. DeBerardinis, Cell Metab., 2011, 14, 285–286 CrossRef CAS PubMed.
- J. W. Locasale, A. R. Grassian, T. Melman, C. A. Lyssiotis, K. R. Mattaini, A. J. Bass, G. Heffron, C. M. Metallo, T. Muranen, H. Sharfi, A. T. Sasaki, D. Anastasiou, E. Mullarky, N. I. Vokes, M. Sasaki, R. Beroukhim, G. Stephanopoulos, A. H. Ligon, M. Meyerson, A. L. Richardson, L. Chin, G. Wagner, J. M. Asara, J. S. Brugge, L. C. Cantley and M. G. Vander Heiden, Nat. Genet., 2011, 43, 869–874 CrossRef CAS PubMed.
- R. Possemato, K. M. Marks, Y. D. Shaul, M. E. Pacold, D. Kim, K. Birsoy, S. Sethumadhavan, H. K. Woo, H. G. Jang, A. K. Jha, W. W. Chen, F. G. Barrett, N. Stransky, Z. Y. Tsun, G. S. Cowley, J. Barretina, N. Y. Kalaany, P. P. Hsu, K. Ottina, A. M. Chan, B. Yuan, L. A. Garraway, D. E. Root, M. Mino-Kenudson, E. F. Brachtel, E. M. Driggers and D. M. Sabatini, Nature, 2011, 476, 346–350 CrossRef CAS PubMed.
- C. F. Labuschagne, N. J. van den Broek, G. M. Mackay, K. H. Vousden and O. D. Maddocks, Cell Rep, 2014, 7, 1248–1258 CrossRef CAS PubMed.
- A. S. Tibbetts and D. R. Appling, Annu. Rev. Nutr., 2010, 30, 57–81 CrossRef CAS PubMed.
- J. W. Locasale, Nat. Rev. Cancer, 2013, 13, 572–583 CrossRef CAS PubMed.
- J. Ye, A. Mancuso, X. Tong, P. S. Ward, J. Fan, J. D. Rabinowitz and C. B. Thompson, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 6904–6909 CrossRef CAS PubMed.
- B. Chaneton, P. Hillmann, L. Zheng, A. C. Martin, O. D. Maddocks, A. Chokkathukalam, J. E. Coyle, A. Jankevics, F. P. Holding, K. H. Vousden, C. Frezza, M. O'Reilly and E. Gottlieb, Nature, 2012, 491, 458–462 CrossRef CAS PubMed.
- W. Du, P. Jiang, A. Mancuso, A. Stonestrom, M. D. Brewer, A. J. Minn, T. W. Mak, M. Wu and X. Yang, Nat. Cell Biol., 2013, 15, 991–1000 CrossRef CAS PubMed.
- W. Hu, C. Zhang, R. Wu, Y. Sun, A. Levine and Z. Feng, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 7455–7460 CrossRef CAS PubMed.
- S. Suzuki, T. Tanaka, M. V. Poyurovsky, H. Nagano, T. Mayama, S. Ohkubo, M. Lokshin, H. Hosokawa, T. Nakayama, Y. Suzuki, S. Sugano, E. Sato, T. Nagao, K. Yokote, I. Tatsuno and C. Prives, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 7461–7466 CrossRef CAS PubMed.
- A. Giacobbe, L. Bongiorno-Borbone, F. Bernassola, A. Terrinoni, E. K. Markert, A. J. Levine, Z. Feng, M. Agostini, L. Zolla, A. F. Agro, D. A. Notterman, G. Melino and A. Peschiaroli, Cell Cycle, 2013, 12, 1395–1405 CrossRef CAS PubMed.
- T. Velletri, F. Romeo, P. Tucci, A. Peschiaroli, M. Annicchiarico-Petruzzelli, M. V. Niklison-Chirou, I. Amelio, R. A. Knight, T. W. Mak, G. Melino and M. Agostini, Cell Cycle, 2013, 12, 3564–3573 CrossRef CAS PubMed.
- O. D. Maddocks, C. R. Berkers, S. M. Mason, L. Zheng, K. Blyth, E. Gottlieb and K. H. Vousden, Nature, 2013, 493, 542–546 CrossRef CAS PubMed.
- I. Amelio, E. K. Markert, A. Rufini, A. V. Antonov, B. S. Sayan, P. Tucci, M. Agostini, T. C. Mineo, A. J. Levine and G. Melino, Oncogene, 2014, 33, 5039–5046 CrossRef CAS PubMed.
- D. E. Bauer, G. Hatzivassiliou, F. Zhao, C. Andreadis and C. B. Thompson, Oncogene, 2005, 24, 6314–6322 CrossRef CAS PubMed.
- P. Nicklin, P. Bergman, B. Zhang, E. Triantafellow, H. Wang, B. Nyfeler, H. Yang, M. Hild, C. Kung, C. Wilson, V. E. Myer, J. P. MacKeigan, J. A. Porter, Y. K. Wang, L. C. Cantley, P. M. Finan and L. O. Murphy, Cell, 2009, 136, 521–534 CrossRef CAS PubMed.
- C. H. Eng, K. Yu, J. Lucas, E. White and R. T. Abraham, Sci. Signaling, 2010, 3, ra31 Search PubMed.
- M. Meng, S. Chen, T. Lao, D. Liang and N. Sang, Cell Cycle, 2010, 9, 3921–3932 CrossRef CAS.
- M. Yuneva, N. Zamboni, P. Oefner, R. Sachidanandam and Y. Lazebnik, J. Cell Biol., 2007, 178, 93–105 CrossRef CAS PubMed.
- D. R. Wise, R. J. DeBerardinis, A. Mancuso, N. Sayed, X. Y. Zhang, H. K. Pfeiffer, I. Nissim, E. Daikhin, M. Yudkoff, S. B. McMahon and C. B. Thompson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18782–18787 CrossRef CAS PubMed.
- P. Gao, I. Tchernyshyov, T. C. Chang, Y. S. Lee, K. Kita, T. Ochi, K. I. Zeller, A. M. De Marzo, J. E. Van Eyk, J. T. Mendell and C. V. Dang, Nature, 2009, 458, 762–765 CrossRef CAS PubMed.
- D. Rieger and N. M. Loskutoff, J. Reprod. Fertil., 1994, 100, 257–262 CrossRef CAS PubMed.
- R. J. DeBerardinis, A. Mancuso, E. Daikhin, I. Nissim, M. Yudkoff, S. Wehrli and C. B. Thompson, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 19345–19350 CrossRef CAS PubMed.
- J. Neermann and R. Wagner, J. Cell. Physiol., 1996, 166, 152–169 CrossRef CAS.
- M. J. Seltzer, B. D. Bennett, A. D. Joshi, P. Gao, A. G. Thomas, D. V. Ferraris, T. Tsukamoto, C. J. Rojas, B. S. Slusher, J. D. Rabinowitz, C. V. Dang and G. J. Riggins, Cancer Res., 2010, 70, 8981–8987 CrossRef CAS PubMed.
- A. C. Bester, M. Roniger, Y. S. Oren, M. M. Im, D. Sarni, M. Chaoat, A. Bensimon, G. Zamir, D. S. Shewach and B. Kerem, Cell, 2011, 145, 435–446 CrossRef CAS PubMed.
- S. S. Chirala, H. Chang, M. Matzuk, L. Abu-Elheiga, J. Mao, K. Mahon, M. Finegold and S. J. Wakil, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 6358–6363 CrossRef CAS PubMed.
- P. W. Hochachka, Living without oxygen: closed and open systems in hypoxia tolerance, Harvard University Press, Cambridge, 1980 Search PubMed.
- P. S. Ward and C. B. Thompson, Cancer Cell, 2012, 21, 297–308 CrossRef CAS PubMed.
- S. Abu-Hamad, H. Zaid, A. Israelson, E. Nahon and V. Shoshan-Barmatz, J. Biol. Chem., 2008, 283, 13482–13490 CrossRef CAS PubMed.
- N. Majewski, V. Nogueira, P. Bhaskar, P. E. Coy, J. E. Skeen, K. Gottlob, N. S. Chandel, C. B. Thompson, R. B. Robey and N. Hay, Mol. Cell, 2004, 16, 819–830 CrossRef CAS PubMed.
- R. Zalk, A. Israelson, E. S. Garty, H. Azoulay-Zohar and V. Shoshan-Barmatz, Biochem. J., 2005, 386, 73–83 CrossRef CAS PubMed.
- S. Shimizu, M. Narita and Y. Tsujimoto, Nature, 1999, 399, 483–487 CrossRef CAS PubMed.
- Y. Cheng, D. Diao, H. Zhang, Q. Guo, X. Wu, Y. Song and C. Dang, Biomed. Rep., 2014, 2, 188–192 CAS.
- R. W. Wannemacher Jr, J. G. Pace, R. A. Beall, R. E. Dinterman, V. J. Petrella and H. A. Neufeld, J. Clin. Invest., 1979, 64, 1565–1572 CrossRef CAS PubMed.
- A. M. Poff, C. Ari, P. Arnold, T. N. Seyfried and D. P. D'Agostino, Int. J. Cancer, 2014, 135, 1711–1720 CrossRef CAS PubMed.
- S. K. Shukla, T. Gebregiworgis, V. Purohit, N. V. Chaika, V. Gunda, P. Radhakrishnan, K. Mehla, I. I. Pipinos, R. Powers, F. Yu and P. K. Singh, Canc. Metabol., 2014, 2, 18 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2015 |