Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Amino acid-based poly(ester amide) nanofibers for tailored enzymatic degradation prepared by miniemulsion-electrospinning†
S. K.
Murase
ab,
L.-P.
Lv
b,
A.
Kaltbeitzel
b,
K.
Landfester
b,
L. J.
del Valle
a,
R.
Katsarava
c,
J.
Puiggali
*a and
D.
Crespy
*b
aDepartment of Chemical Engineering, ETSEIB, Universitat Politècnica de Catalunya, Av. Diagonal 647, 08028, Barcelona, Spain. E-mail: jordi.puiggali@upc.edu
bMax Planck Institute for Polymer Research, Ackermannweg 10, 55128 Mainz, Germany. E-mail: crespy@mpip-mainz.mpg.de
cInstitute of Chemistry and Molecular Engineering, Agricultural University of Georgia, 13 km David Aghmashenebeli Alley, Tblisi 0159, Georgia
First published on 9th June 2015
Abstract
Novel enzyme loaded scaffolds with enzyme-responsive degradable properties for drug delivery are prepared by an original inverse-miniemulsion electrospinning method. Miniemulsions with aqueous nanodroplets containing different enzymes, i.e. lipase or α-chymotrypsin, and a fluorophore are electrospun with a solution of poly(ester amide) and polycaprolactone to fabricate multicompartment nanofibers. The poly(ester amide) contains the two essential amino acids phenylalanine and leucine that promote low cytotoxicity degradation products and makes them suitable for the preparation of drug delivery devices for the biomedical field. The activity of the loaded enzymes in different conditions and a sustained degradation of fibers mechanism with an approximate 20% weight loss within one month are observed. Locating enzymes in degradation medium accelerated the degradation until complete scaffold destruction in less than 5 days. In all cases, a nearly complete release of the loaded fluorophore (from 80% and upwards) was achieved before the complete degradation of fibers occurred, suggesting that the nanofibers are suitable as self-triggered drug release systems with sustained mechanical integrity and a flexible range of degradation rates.
Introduction
Electrospinning is nowadays a conventional method for the preparation of non-wovens of polymer nano- and microfibers.1,2 Fibers loaded with drugs,3,4 and biological agents, i.e. viruses, bacteria, enzymes,5–7 were extensively investigated due to the advantages associated with protection by encapsulation, controlled release, and ease of preparation that this technique allows for. However, using the electrospinning technique to prepare fibers for biomedical application still presents some challenges that must be overcome. Indeed, the fiber's matrix should be biodegradable and insoluble in water. The polymers used to build the matrix are therefore usually hydrophobic and have to be usually dissolved in organic solvents for the formulation. However, biomolecules tend to be very sensitive to organic solvents and are easily denatured during the preparation process. A potential candidate to overcome these drawbacks is coaxial electrospinning, which is based on the use of a dual-capillary spinneret that maintains inner and outer solutions separated.8 However, the process cannot be applied to every material. Colloid-electrospinning is a relatively novel technique that has attracted growing interest for the encapsulation of bioactive agents.9–12 It offers the possibility to produce tailorable nanofibers with different components and nanostructures. A polymer solution is mixed with a colloidal suspension with the purpose of incorporating a second functional substance into the fibers without the requirement of further post-treatments.13–15 The use of water-in-oil (W/O) emulsions allows the introduction of selected hydrophilic bioactive substances in the aqueous phase that will be finally located in the fibers. Examples of successfully encapsulated hydrophilic substances are small drugs,13,16 proteins,17,18 enzymes,19 and the combination of two drugs.20,21 Biological molecules were not affected by the presence of organic solvents because they were emulsified, so that the activity of the biological agents was maintained after the preparation method. However, the presence of colloids and the resulting behavior of the polymer solutions increase the complexity of the different parameters involved in the process.In all the previous examples, only the most common biodegradable polyesters, i.e. polylactide, polyglycolide, or polycaprolactone (PCL) have been used. PCL is a commonly reported biodegradable polymer for different medical applications.22,23 However, the degradation of polyesters usually presents a fast kinetic degradation rate performed by hydrolytic or enzymatic attack of the ester group, meaning that the mechanical stability also tends to decrease accordingly. The inclusion of amide units in the biodegradable material can increase the mechanical and thermal properties due to the presence of hydrogen bonding between these units and include a new cleavable position. Although mixtures of PEA and PCL were already reported in the biomedical field,24 they have never been employed to prepare core–sheath fibers by the colloid-electrospinning method. For this purpose, the copolymer coPEA 8-[L-Phe-6]0.95–[L-Leu-6]0.05 (Fig. S1†), will be used in this study. It incorporates the two amino acids phenylalanine and leucine in its main chain, resulting in two enzymatically cleavable bonds that can be selectively attacked by specific enzymes for enhancing its biodegradability.25
The prepared scaffold should then provide a higher control of degradability and release of the loaded drugs. This tuneable degradation process is convenient to mimic the different regeneration processes in the body. For example, coPEA scaffolds can be used as temporary surgical devices for tissue regeneration.26 Furthermore, the presence of these natural amino acids in the main chain increase the bioassimilation of the resulting degraded products compared to the classical synthetic biopolymers.27 Scaffolds prepared by colloid-electrospinning are ideal candidates for enzyme immobilization because of their high surface-to-volume ratio, porous morphology, and biodegradability.
Exploiting the presence of enzymes in nanomaterials renders a wide range of smart applications based in the modulation of drug delivery profiles or triggering drug release thanks to their exceptional selectivity and specificity.28 Encapsulated enzymes can become active at certain targeted sites enhancing drug release specificity and also lower side-effects. In addition, they can be used for signalling physiological changes or site-targeted drug delivery.29 Examples of stimuli-responsive polymers can be found in several fields, such as anticancer therapeutic systems,30 polymeric amphiphiles,31 gastro-intestinal tract (GIT), or colon-targeted drug delivery.29,32
The objective of the present paper is to demonstrate the feasibility of incorporating active biomolecules such as enzymes and fluorescent labels in the fibers to determine the release behavior of hydrophilic drugs under rapid enzymatic degradation of the matrix. For such purpose, a blend of PCL and coPEA was used to study the degradation mechanism of the prepared scaffolds. In addition, based on previous studies on amino acid-based PEAs and PCL,33–38 a non-specific lipase that catalyses the hydrolysis of esters and α-chymotrypsin, a specific serine protease, were selected to determine the enzyme-responsive performance of the designed devices.
Experimental section
Materials
Lipase from Pseudomonas cepacia (Sigma-Aldrich), α-chymotrypsin from bovine pancreas (Sigma-Aldrich), Rhodamine 6G chloride (R6G, Sigma-Aldrich, 99%), Lubrizol OS 210980 (Lubrizol Co.) and polycaprolactone (PCL, Mw = 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–90
000–90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1, Sigma-Aldrich) were used as received. The copoly(ester amide) coPEA 8-[L-Phe-6]0.95–[L-Leu-6]0.05 was synthesized as reported previously (Mw = 42
000 g mol−1, Sigma-Aldrich) were used as received. The copoly(ester amide) coPEA 8-[L-Phe-6]0.95–[L-Leu-6]0.05 was synthesized as reported previously (Mw = 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) (Fig. S1†).39 1× DPBS (Gibco®) buffer without calcium or magnesium and pH 7.4 was used in the different assays. The antimicrobial agent sodium azide (0.02% w/v) and calcium chloride (5 mM) as a cofactor were added to the buffer.
000 g mol−1) (Fig. S1†).39 1× DPBS (Gibco®) buffer without calcium or magnesium and pH 7.4 was used in the different assays. The antimicrobial agent sodium azide (0.02% w/v) and calcium chloride (5 mM) as a cofactor were added to the buffer.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v
1, v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) or dichloromethane/cyclohexane/formic acid (1
v) or dichloromethane/cyclohexane/formic acid (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.075, v
0.075, v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v
v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v), depending on the final scaffold, and 15 mg of Lubrizol. The specific quantities are shown in Table 1. The mixture was stirred at 1000 rpm for 1 h and ultrasonicated with a Branson sonifier ultrasound (W-450D, 1/2 inch tip) at 90% amplitude for 2 min (30 s pulse to 10 s pause). DLS measurements were performed at 5–30–60 min to control the stability of the droplets (Table 1). The scheme for the preparation is displayed in Fig. 1a.
v), depending on the final scaffold, and 15 mg of Lubrizol. The specific quantities are shown in Table 1. The mixture was stirred at 1000 rpm for 1 h and ultrasonicated with a Branson sonifier ultrasound (W-450D, 1/2 inch tip) at 90% amplitude for 2 min (30 s pulse to 10 s pause). DLS measurements were performed at 5–30–60 min to control the stability of the droplets (Table 1). The scheme for the preparation is displayed in Fig. 1a.
| Continuous phase (14 mL) | Dispersed phase (10 mL) | DLS | ||||||
|---|---|---|---|---|---|---|---|---|
| Lubrizol [mg] | Mixturea | NaCl [mg mL−1 water] | Dye [mg] | Enzymeb [mg g−1 polymer] | D h [nm] | σ (%) | ||
| Ratio | ||||||||
a M1: DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C6H12, M2: DCM C6H12, M2: DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C6H12 C6H12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HCOOH.
b Lipase and α-chymotrypsin named L and CT, respectively. HCOOH.
b Lipase and α-chymotrypsin named L and CT, respectively.
|
||||||||
| 15 | M1 | (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
25 | 0 | No | 0 | 320 | 42 |
| 15 | M2 | (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.075) 0.075) |
25 | 0 | No | 0 | 640 | 48 |
| 15 | M1 | (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
25 | 0.5 | No | 0 | 400 | 44 |
| 15 | M2 | (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.075) 0.075) |
25 | 0.5 | No | 0 | 400 | 30 |
| 15 | M2 | (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.075) 0.075) |
25 | 0.5 | L | 15 | 310 | 41 |
| 15 | M2 | (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.075) 0.075) |
25 | 0.5 | CT | 15 | 490 | 36 |
| Polymer | Mixtureb | Enzymec [mg g−1 polymer] | Flow rate [mL h−1] | Voltage [kV] | ||||
|---|---|---|---|---|---|---|---|---|
| Namea | Composition | % [g mL−1 solution] | Ratio | |||||
a Polymer composition as PCL or polymer mixture (PM) PCL/coPEA 70/30 and the subscript indicating the loaded enzyme L or CT as lipase or α-chymotrypsin, respectively.
b M1: DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C6H12, M2: DCM C6H12, M2: DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C6H12 C6H12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HCOOH.
c Lipase and α-chymotrypsin named L and CT, respectively. HCOOH.
c Lipase and α-chymotrypsin named L and CT, respectively.
|
||||||||
| PCL | PCL | 10 | M1 | (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
No | 0 | 0.5 | 15 |
| PCLL | PCL | 10 | M1 | (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) 1) |
L | 15 | 0.85 | 12 |
| PM | PCL/coPEA 70/30 | 12 | M2 | (17.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.3 4.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) 3) |
No | 0 | 5 | 15 |
| PML | PCL/coPEA 70/30 | 12 | M2 | (17.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.3 4.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
L | 10 | 6 | 15 |
| PMCT | PCL/coPEA 70/30 | 10 | M2 | (17.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.3 4.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) 5) |
CT | 10 | 5 | 15 |
| Name | Polymer composition | Loaded enzyme | Medium |
|---|---|---|---|
| PCL | PCL | No | DPBS |
| PCLL | PCL | Lipase | DPBS |
| PM | PCL/coPEA 70/30 | No | DPBS |
| PM-L | PCL/coPEA 70/30 | No | DPBS + lipase |
| PM-CT | PCL/coPEA 70/30 | No | DPBS + chymotrypsin |
| PML | PCL/coPEA 70/30 | Lipase | DPBS |
| PML-CT | PCL/coPEA 70/30 | Lipase | DPBS + α-chymotrypsin |
| PMCT | PCL/coPEA 70/30 | α-Chymotrypsin | DPBS |
| PMCT-L | PCL/coPEA 70/30 | α-Chymotrypsin | DPBS + lipase |
Analytical methods
The hydrodynamic diameters (Dh) of the aqueous droplets were estimated with a Nicomp 380 Submicron Particle Sizer (PSS-Nicomp) at room temperature and a laser diode wavelength of 635 nm. The measurements were performed in triplicate at 5, 30, and 60 min and are reported as the average value at 60 min. The morphology of the fibers was examined with a Leo Gemini 1530 (Carl Zeiss AG, Oberkochem, Germany) scanning electron microscope (SEM) operating at 0.1–0.2 kV. 0.5 × 0.5 cm pieces of aluminium foil covered with the different electrospun fibers were observed. Confocal laser scanning microscope (CLSM) images were obtained using a Leica TCS SP5 (Leica Mikrosysteme Vertrieb GmbH, Wetzlar) microscope equipped with a Leica HCX PL APO CS 1.4/63× objective. Rhodamine 6G chloride (0.1 M or 0.05 g mL−1), a water-soluble fluorescent dye, was loaded in the aqueous droplets to determine their distribution in the fibers. The fluorophore has a λmax(absorption) = 526 nm and a λmax(emission) = 555 nm. The setup for colloid-electrospinning was previously described.9 A commercial electrospin cabin dual voltage from IME Technologies was used. A disposable syringe loaded with the electrospinning suspension was placed in a Harvard apparatus PHD 2000 syringe pump and connected to a metal tip end (ID: 0.5 mm) with plastic tubing. The concentration of the fluorophore released from the fibers was determined with a plate reader Infinite M1000 (Tecan, Germany). Calibration curves of the dye in the different media including enzymes were prepared. The 96-well black tray was fluorescent top read with λ(excitation) = 526 nm and λ(emission) = 556 nm at 25 °C.Results and discussion
Preparation of the scaffolds
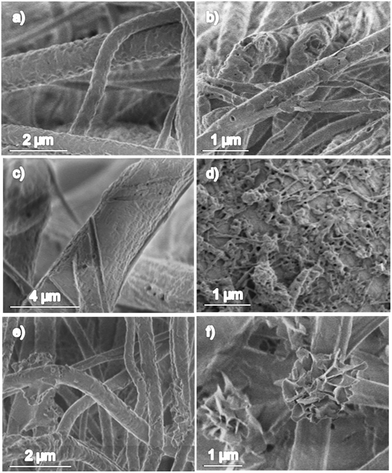
Different miniemulsions were prepared to maximize the stability of the inverse miniemulsions in the presence of the polymers dissolved in the continuous phase. NaCl was included in the aqueous dispersed phase as lipophobe, whose role is to increase the osmotic pressure inside the droplets and suppress the Ostwald ripening effect.40 A concentration of 25% (w/v) of NaCl in the miniemulsion was the minimum needed to obtain stable aqueous droplets in the different emulsions. The addition of very low amount of formic acid in the miniemulsion was required to completely dissolve the copolymer. This concentration did not affect the activity of the loaded enzymes.In the electrospinning process, the variation of the flow rate, electric field, tip-collector distance, viscosity of the solution, temperature, and relative humidity results in fibers with different properties and morphologies.41,42 To form the PCL fibers, a minimum concentration of 10% (w/v) of polymer was needed, whereas for the mixture PCL/coPEA 70/30 an increase to 12% (w/v) was required for samples PM and PML (Table 2) due to the lower molecular weight of the coPEA. However, higher concentrations (40–50% w/v) led to inhomogeneous and discontinuous fibers. The feed rate was adjusted to be below or equal to 1 mL h−1 (0.5 and 0.85 mL h−1 for samples PCL and PCLL, respectively, Table 2). For higher rates, the fiber diameter was thinner and beads were observed along the fibers (Fig. S2a†). The latter phenomena, already been described for other polymers such as viscose,43 PS,44 chitosan/PLA mixtures45 or PMMA,46 appears when there is an uneven evaporation of the solvent between the skin and the core, creating a dry external layer before the internal solvent completely evaporates and finally causing the jet to collapse. Increasing the flow rate resulted in a more pronounced effect, creating completely flat and empty tubes with holes all over the surface, as seen in Fig. S2b.† Due to the low glass transition temperature of the coPEA (Tg ∼ 26 °C), the coPEA fibers did not conserve their structural integrity at room temperature (Fig. S3a†). The fibers observed by CLSM revealed homogeneous fluorescence inside the fibers, indicating that the structure of the aqueous droplets were not maintained in the fibers as well as no formation of core–sheath morphology (see Fig. S3b†). Therefore, the coPEA was mixed with PCL with a ratio of PCL/coPEA of 70/30 to obtain fibers with cylindrical cross-sections and with aqueous compartments dispersed inside the fibers (samples PM, PML and PMCT, Table 2). Flat PCL/coPEA fibers were obtained when lipase was introduced in the mixture (sample PML in Table 3 and Fig. S2c†) due to the aforementioned uneven evaporation event. In addition, when the flow rate was higher than 6 mL h−1, holes could be detected along the centre of the flat surface, similar to the ones previously described for PCL fibers (Fig. S2b†). The α-chymotrypsin enzyme could also be efficiently encapsulated in the PCL/coPEA fibers (Table 4 and Fig. S2d†). The average value of the diameters of the prepared fibers is shown in Table 4 and the diameter distribution can be seen in Fig. S4.† When PCL fibers were loaded with lipase, the diameter showed a drastic decrease of 50% from 744 to 375 nm (PCL and PCLL in Table 3, Fig. 2b and c). The slight increase in flow rate (from 0.5 for sample PCL to 0.85 mL h−1 for sample PCLL) needed to prepare continuous enzyme-loaded PCL scaffolds (sample PCLL) caused an expected reduction in the resulting diameter. It is also noteworthy to mention that fibers that included the coPEA exhibited a larger diameter in comparison with pure PCL fibers. This was due to the low Tg of the PEA resulting in the flattening of the fibers (Fig. S4b†). Fibers containing coPEA displayed a flat ribbon-like or wrinkled surface due to phase-separation (Fig. 2d). In order to localize the aqueous compartments in the fibers, we observed them by fluorescence microscopy because it is a method particularly suitable for identifying nanostructures in polymer nanofibers.47,48 The fibers displayed some fluorescence, which can be observed in the CLSM images of Fig. 3 in dark blue color. The light green color indicates a higher concentration of dye. The fluorescent domains confirm the presence and distribution of R6G in reservoirs inside the fibers. It is assumed that the enzymes were co-localized with the dye since both were present in the dispersed phase of the inverse miniemulsion.
| Sample | Fiber diameter | |
|---|---|---|
| Average [nm] | σ [%] | |
| PCL | 745 | 21 |
| PCLL | 375 | 27 |
| PM | 1377 | 27 |
| PML | 1000 | 44 |
| PMCT | 966 | 26 |
 | ||
| Fig. 2 SEM micrographs of the prepared electrospinning scaffolds. (a) PCL without R6G, (b) PCL, (c) PCLL, (d) PM, (e) PML, and (f) PMCT. See nomenclature in Table 2. Insets show a magnification of the respective samples. | ||
 | ||
| Fig. 3 CLSM images of (a) sample PCL, (b and c) PM, and (d) PMCT. The scale bar represents 25 μm or 1 μm in (c). | ||
Enzymatic degradation assays
Enzymatic degradation of PCL with different morphologies, i.e., films,34,49 particles,36 electrospun mats,35 and blends50 were extensively studied with lipases. Lipase cleaves non-specific ester linkages so an enzymatic degradation of PCL is expected. Results of the degradation behavior exhibited by lipase loaded PCL scaffolds (named PCLL) are shown in Fig. 4a. PCL scaffolds loaded with the enzyme and placed in DPBS medium for 31 days featured a noticeable decrease of nearly 10% of their initial weight in the first 100 h. Then, the diffusion of the medium in the fibers took place and the loaded enzymes could be activated to start the degradation process from inside of the fibers. However, the degradation process attained a linear behavior after 200 h, suggesting an accused slowdown in the enzymatic activity. PCL is relatively hydrophobic,51 which may hinder a complete penetration of the medium into internal layers. Besides, semicrystalline polymers such as PCL tend to present a first fast degradation stage related to medium diffusion and breakage of amorphous regions, followed by a second slow stage related to the degradation of the crystalline regions. | ||
| Fig. 4 Weight loss of the nanofibers by enzymatic degradation versus time for (a) PCL and PCLL scaffolds and (b) PCL/coPEA 70/30 scaffolds (PM samples). | ||
In Fig. 2c, a SEM image of PCL scaffolds loaded with lipase (sample PCLL) can be seen. And in Fig. 5b, a recovered sample after 198 h of immersion in DPBS medium is shown. The surface of the fibers display a series of pores and peeling in fine layers that were not present at the beginning of the assay due to the action of the enzymes. To determine the stability of the PCL scaffold in medium and the possible hydrolytic effect, sample PCL (scaffold without enzyme) was placed in DPBS buffer. Only 1% of weight was lost in 33 days remaining steady afterwards, which ascertains a negligible effect of the medium on the scaffold, in accordance to previous studies on PCL.33 The lost weight in sample PCLL (PCL with loaded lipase) can be attributed to the enzymatic activity and the resulting surface morphology in Fig. 5a.
Degradation assays were also carried out in two enzymatic media with scaffolds prepared with the mixture of PCL/coPEA 70/30 named PM (see Table 3). To determine the hydrolytic effect of the buffer on these samples, mats of sample PM were introduced in DPBS buffer without enzyme, resulting in an only 4% weight loss in 1 month. The immersed scaffolds experienced a negligible weight loss with comparison to the effect of the enzymes. The same scaffolds without enzyme (sample PM) were introduced in DPBS enzymatic media containing α-chymotrypsin (PM-CT), losing a significant weight percentage in 4 days, 60% (Fig. 4b). Inclusion of the enzymes into the fibers decreased considerably the degradation advancement, as can be seen for samples PML and PMCT in Fig. 4 (nomenclature in Table 4). After 6 days, the lipase-loaded fiber (sample PML) lost 10% weight while the α-chymotrypsin-loaded fiber (sample PMCT), an 18%. Diffusion and activation of the enzyme were both hindered by the polymer hydrophobicity, clearly delaying the degradation process. The difference between the degradation rates between samples PML and PMCT may be associated to two effects. Firstly, the more flat ribbon-like morphology of sample PML and more cylindrical for the latter. Secondly, due to the specificity of α-chymotrypsin towards amide groups found between large hydrophobic amino acids, in this case phenylalanine in coPEA.
When enzyme-loaded scaffolds (PML and PMCT) are introduced in enzymatic media (sample PML-CT and PMCT-L, see Table 3), two degradation steps (Fig. 4b) that are associated to two different effects were observed. The first step (first 80 h) can be associated to the degradation induced by the enzymes present in the media (α-chymotrypsin for PML-CT and lipase for PMCT-L), exhibiting a very similar behavior to sample PM-CT, for which the enzyme is only in the external medium. The sample PMCT-L in the lipase medium lost 42% of its initial weight while sample PML-CT in the α-chymotrypsin medium lost 54% after 80 h. Sample PML-CT immersed in the α-chymotrypsin medium was degraded faster than the one with lipase in the medium, although the scaffolds were also loaded with enzymes, suggesting a more effective attack by α-chymotrypsin when it is placed in the outer degradation medium. In the second step, the degradation promoted by the enzymes inside the fibers also took place. The difference in the degradation rate between the two samples PML-CT and PMCT-L was more pronounced in this phase (20% difference). This deviation can be associated to a partial proteolytic digestion of lipase by α-chymotrypsin, which resulted in a slowdown of the degradation rate of α-chymotrypsin loaded fibers (PMCT-L). In spite of the partial digestion effect, the loaded enzymes maintained their activity after the electrospinning process as shown in Fig. 4, demonstrating that the colloid electrospinning technique has a potential use in the preparation of scaffolds loaded with hydrophilic biological molecules. Loading the enzymes in the fibers and also adding them to the buffer medium (samples PML-CT and PMCT-L) did not double their activity, but allowed for an intermediate degradation with more than one step, which can be tuned varying the quantity and type of enzyme. Remarkably, the difference in the degradation rate between sample PM-CT (in α-chymotrypsin media) and sample PML-CT (lipase loaded in α-chymotrypsin media) was about 7% in the first 80 h and 11% between PM-CT and PMCT-L (α-chymotrypsin loaded in lipase media). The effectiveness of α-chymotrypsin in the outer medium that governs the first step yielded a similar behavior for PM-CT and PML-CT. Both samples were degraded at faster rates than sample PMCT-L. In the second step, the activation of the loaded lipase in sample PML-CT accelerated the overall weight loss, boosting the second degradation step. This is a consequence of the presence of α-chymotrypsin in the outer media that increased the diffusion of the medium into the fiber. A similar enhancement in degradation rate was also observed for sample PMCT-L, but still with a global slower degradation rate.
The resulting morphologies of degraded fibers at certain times are shown in Fig. 5 and also compared to the non-degraded fibers (Fig. 2). A different morphology appeared on the surface of sample PMCT-L (see Fig. 5f (ref. 52)). The lipase present in the medium of this experiment formed nanostructures on different areas of the scaffold. The deposition of the enzyme on the surface of the fibers did not cleave the polymer, and consequently slowed down the degradation rate.
Release of loaded fluorophore
PCL and coPEA displayed different degradation behaviors controlled by the type and localization of the enzymes. Therefore, the release behavior for the dye loaded in the fibers should be influenced by factors related to the matrix (composition and degradation), release media (DPBS or enzymatic media), and the loaded drug (interaction with matrix and with medium). The release kinetics obtained by fluorescence spectroscopy from the samples (PM-CT, PML-CT and PMCT-L, Table 3) can be observed in Fig. 6. All samples exhibited two differentiated steps: the first one for the initial 10 h followed by a zero-order or a pseudo-first order phase. A burst effect can be also observed in all samples that delivered between an 11 and a 17% of dye in the first 15 min, showing sample PML-CT the most pronounced burst. Among the diverse mathematical models presented by Tzafriri,52 one describes the different steps of the obtained results. The model, usually used for polyester devices,53 predicts an initial step driven by diffusion and a second step governed by the degradation process. In this case, using a PCL/coPEA mixture of 70/30, it was expected that the polyester guided the final release profile. Assuming that there was negligible degradation of the matrix in the first hours, the release of the loaded dye to the outer medium was accomplished by Fickian diffusion, a first order t1/2-time dependent model and driven by the concentration gradient of the fluorophore.54 Dye release from flat ribbon-like samples (PML-CT) has a slightly different release profile than from cylindrical samples (PM-CT and PMCT-L). Sample PML-CT can be approached by a zero-order model with a burst effect55 and PM-CT and PMCT-L samples show a first order model release behavior. However, taking into account the previous degradation results, some samples underwent degradation from the beginning of the release experience, adding a degradation factor in the release mechanism. It is then evidenced that the prepared samples that contain coPEA (sample PM-CT, PML-CT and PMCT-L) have a first step mainly governed by diffusion but that can be modified at will by the degradation factor, resulting in a combined profile. | ||
| Fig. 6 R6G release percentage determined by fluorescent detection versus time for the different scaffolds (PM) in enzymatic media (L: lipase and CT: α-chymotrypsin). | ||
If we focus on the second release step, samples PM-CT and PMCT-L, displayed the aforementioned pseudo-first order phase, attaining an 80% release of the loaded dye at 120 h (5 days). Sample PML-CT exhibited a zero-order mechanism that arrives to a complete release at the end of the 120 h. In terms of degradation of the scaffold PML-CT is not completely degraded at 120 h. This suggests that the diffusion phenomenon was the driving force for the dye release and not the degradation of the fiber, what also happened for sample PM-CT and PMCT-L. In addition, the slower release profile of sample PML-CT, could be attributed to the flat ribbon-like morphology of that sample (see Fig. 2e) and the disposition of the internal nanoreservoirs. A saturated dye solution was created at the core of the fibers due to the dissolution of the dye into the diffused media. The diffusion of this solution towards the external media was slower than this internal dye dissolution, creating an internal constant dye excess and giving rise to the zero-order release profile.55 Besides, the aforementioned partial digestion of the loaded lipase also affected the release of the dye. The release process was decelerated until diffusion of the medium activated the internal lipase and overcame the proteolysis.
Conclusions
Enzymes and a fluorescent dye were successfully loaded and distributed as nanoreservoirs into fibers by electrospinning inverse miniemulsions. The encapsulation and remaining activity of the loaded enzymes and fluorescent dye were proven by the subsequent degradation and release assays. The resulting degradation kinetics revealed different behaviors depending on the position and type of enzyme. α-Chymotrypsin degraded the scaffold faster when placed in the external medium, and also when it was loaded in the fibers. Placing enzyme-loaded scaffolds in enzymatic media gave rise to a two-steps degradation profile. A first degradation step governed by the attack of the external enzymes was observed, followed by a faster degradation rate in the second step due to activation of the internal enzymes. At the beginning of the experiment, the degradation mechanism of the loaded fibers was 5–10% slower with respect to the unloaded ones, which is explained by the partial proteolysis between the two enzymes. The use of a blend of polycaprolactone and polyester amide and the combination of enzymes present inside the fibers or/and in the external medium allow for the preparation of therapeutic scaffolds that respond to different triggers, yielding therefore a large range of biodegradation rates. Such properties enabled a controlled release of a payload that was co-encapsulated in the nanoreservoirs present in the fibers showing the suitability of the prepared device as an enzyme-responsive material for drug-delivery.Acknowledgements
S.K.M. acknowledges an EEBB-FPI grant from MICINN.References
- Z.-M. Huang, Y.-Z. Zhang, M. Kotaki and S. Ramakrishna, Compos. Sci. Technol., 2003, 63, 2223–2253 CrossRef CAS.
- D. Li and Y. Xia, Adv. Mater., 2004, 16, 1151–1170 CrossRef CAS PubMed.
- M. Hamori, S. Yoshimatsu, Y. Hukuchi, Y. Shimizu, K. Fukushima, N. Sugioka, A. Nishimura and N. Shibata, Int. J. Pharm., 2014, 464, 243–251 CrossRef CAS PubMed.
- X. Hu, S. Liu, G. Zhou, Y. Huang, Z. Xie and X. Jing, J. Controlled Release, 2014, 185, 12–21 CrossRef CAS PubMed.
- P. Yilgor, R. A. Sousa, R. L. Reis, N. Hasirci and V. Hasirci, J. Mater. Sci.: Mater. Med., 2010, 21, 2999–3008 CrossRef CAS PubMed.
- W. Salalha, J. Kuhn, Y. Dror and E. Zussman, Nanotechnology, 2006, 17, 4675–4681 CrossRef CAS PubMed.
- Y. Dror, J. Kuhn, R. Avrahami and E. Zussman, Macromolecules, 2008, 41, 4187–4192 CrossRef CAS.
- Z. Sun, E. Zussman, A. L. Yarin, J. H. Wendorff and A. Greiner, Adv. Mater., 2003, 15, 1929–1932 CrossRef CAS PubMed.
- D. Crespy, K. Friedemann and A.-M. Popa, Macromol. Rapid Commun., 2012, 33, 1978–1995 CrossRef CAS PubMed.
- E. Jo, S. Lee, K. T. Kim, Y. S. Won, H.-S. Kim, E. C. Cho and U. Jeong, Adv. Mater., 2009, 21, 968–972 CrossRef CAS PubMed.
- J. C. Sy, A. S. Klemm and V. P. Shastri, Adv. Mater., 2009, 21, 1814–1819 CrossRef CAS PubMed.
- M. Bannwarth and D. Crespy, Chem.–Asian J., 2014, 9, 2030–2035 CrossRef CAS PubMed.
- X. Xu, L. Yang, X. Xu, X. Wang, X. Chen, Q. Liang, J. Zeng and X. Jing, J. Controlled Release, 2005, 108, 33–42 CrossRef CAS PubMed.
- Y. Dai, J. Niu, J. Liu, L. Yin and J. Xu, Bioresour. Technol., 2010, 101, 8942–8947 CrossRef CAS PubMed.
- K. Wei, Y. Li, X. Lei, H. Yang, A. Teramoto, J. Yao, K. Abe and F. K. Ko, Macromol. Biosci., 2011, 11, 1526–1536 CAS.
- X. Xu, X. Chen, P. Ma, X. Wang and X. Jing, Eur. J. Pharm. Biopharm., 2008, 70, 165–170 CrossRef CAS PubMed.
- Y. Yang, X. Li, W. Cui, S. Zhou, R. Tan and C. Wang, J. Biomed. Mater. Res., Part A, 2008, 86, 374–385 CrossRef PubMed.
- X. Li, Y. Su, S. Liu, L. Tan, X. Mo and S. Ramakrishna, Colloids Surf., B, 2010, 75, 418–424 CrossRef CAS PubMed.
- X. Li, H. Zhang, H. Li and X. Yuan, Colloid Polym. Sci., 2010, 288, 1113–1119 CAS.
- B. Song, C. Wu and J. Chang, Acta Biomater., 2012, 8, 1901–1907 CrossRef CAS PubMed.
- S. Yan, L. Xiaoqiang, L. Shuiping, M. Xiumei and S. Ramakrishna, Colloids Surf., B, 2009, 73, 376–381 CrossRef PubMed.
- M. A. Woodruff and D. W. Hutmacher, Prog. Polym. Sci., 2010, 35, 1217–1256 CrossRef CAS PubMed.
- J. R. Venugopal, Y. Zhang and S. Ramakrishna, Artif. Organs, 2006, 30, 440–446 CrossRef CAS PubMed.
- M. Yasin and B. J. Tighe, Biomaterials, 1992, 13, 9–16 CrossRef CAS.
- Y. Fan, M. Kobayashi and H. Kise, J. Polym. Sci., Part A: Polym. Chem., 2002, 40, 385–392 CrossRef CAS PubMed.
- L. M. Pratt and C. C. Chu, J. Polym. Sci., Part A: Polym. Chem., 1994, 32, 949–960 CrossRef CAS PubMed.
- T. Kartvelishvili, G. Tsitlanadze, L. Edilashvili, N. Japaridze and R. Katsarava, Macromol. Chem. Phys., 1997, 198, 1921–1932 CrossRef CAS PubMed.
- Q. Hu, P. S. Katti and Z. Gu, Nanoscale, 2014, 6, 12273–12286 RSC.
- P. Bawa, V. Pillay, Y. E. Choonara and L. C. du Toit, Biomed. Mater., 2009, 4, 022001 CrossRef PubMed.
- D. B. Pacardo, F. S. Ligler and Z. Gu, Nanoscale, 2015, 7, 3381–3391 RSC.
- C. Wang, Q. Chen, Z. Wang and X. Zhang, Angew. Chem., Int. Ed. Engl., 2010, 49, 8612–8615 CrossRef CAS PubMed.
- L. Yang, J. S. Chu and J. A. Fix, Int. J. Pharm., 2002, 235, 1–15 CrossRef CAS.
- C. X. F. Lam, D. W. Hutmacher, J.-T. Schantz, M. A. Woodruff and S. H. Teoh, J. Biomed. Mater. Res., Part A, 2009, 90, 906–919 CrossRef PubMed.
- Z. Gan, Q. Liang, J. Zhang and X. Jing, Polym. Degrad. Stab., 1997, 56, 209–213 CrossRef CAS.
- H. Peng, J. Ling, J. Liu, N. Zhu, X. Ni and Z. Shen, Polym. Degrad. Stab., 2010, 95, 643–650 CrossRef CAS PubMed.
- D. R. Chen, J. Z. Bei and S. G. Wang, Polym. Degrad. Stab., 2000, 67, 455–459 CrossRef CAS.
- G. Tsitlanadze, M. Machaidze, T. Kviria, N. Djavakhishvili, C. C. Chu and R. Katsarava, J. Biomater. Sci., Polym. Ed., 2004, 15, 1–24 CrossRef CAS PubMed.
- G. Tsitlanadze, T. Kviria, R. Katsarava and C. C. Chu, J. Mater. Sci.: Mater. Med., 2004, 15, 185–190 CrossRef CAS.
- R. Katsarava, V. Beridze, N. Arabuli, D. Kharadze, C. C. Chu and C. Y. Won, J. Polym. Sci., Part A: Polym. Chem., 1999, 37, 391–407 CrossRef CAS.
- D. Crespy, M. Stark, C. Hoffmann-Richter, U. Ziener and K. Landfester, Macromolecules, 2007, 40, 3122–3135 CrossRef CAS.
- G. C. Rutledge and S. V. Fridrikh, Adv. Drug Delivery Rev., 2007, 59, 1384–1391 CrossRef CAS PubMed.
- A. Rogina, Appl. Surf. Sci., 2014, 296, 221–230 CrossRef CAS PubMed.
- R. J. E. Cumberbirch, J. E. Ford and R. E. Gee, J. Text. Inst., Trans., 1961, 52, T330–T350 CrossRef CAS PubMed.
- S. Koombhongse, W. Liu and D. H. Reneker, J. Polym. Sci., Part B: Polym. Phys., 2001, 39, 2598–2606 CrossRef CAS PubMed.
- S.-H. Chen, Y. Chang, K.-R. Lee and J.-Y. Lai, J. Membr. Sci., 2014, 450, 224–234 CrossRef CAS PubMed.
- L. Li, Z. Jiang, M. Li, R. Li and T. Fang, RSC Adv., 2014, 4, 52973–52985 RSC.
- K. Friedemann, A. Turshatov, K. Landfester and D. Crespy, Langmuir, 2011, 27, 7132–7139 CrossRef CAS PubMed.
- C. Herrmann, A. Turshatov and D. Crespy, ACS Macro Lett., 2012, 1, 907–909 CrossRef CAS.
- L. Liu, S. Li, H. Garreau and M. Vert, Biomacromolecules, 2000, 1, 350–359 CrossRef CAS.
- H. Tsuji, Y. Kidokoro and M. Mochizuki, Macromol. Mater. Eng., 2006, 291, 1245–1254 CrossRef CAS PubMed.
- H. Jahani, F. A. Jalilian, C.-Y. Wu, S. Kaviani, M. Soleimani, N. Abassi, K.-L. Ou and H. Hosseinkhani, J. Biomed. Mater. Res., Part A, 2014, 103, 1875–1881 CrossRef PubMed.
- A. R. Tzafriri, J. Controlled Release, 2000, 63, 69–79 CrossRef CAS.
- Y. Fu and W. J. Kao, Expert Opin. Drug Delivery, 2010, 7, 429–444 CrossRef CAS PubMed.
- J. Siepmann and N. A. Peppas, Int. J. Pharm., 2011, 418, 6–12 CrossRef CAS PubMed.
- J. Siepmann and F. Siepmann, J. Controlled Release, 2012, 161, 351–362 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra06267e |
| This journal is © The Royal Society of Chemistry 2015 |