A Geobacter strain isolated from rice paddy soil with higher bioelectricity generation capability in comparison to Geobacter sulfurreducens PCA†
Dandan Deng,
Yichi Zhang and
Ying Liu*
College of Life Sciences, Northwest A&F University, No. 22 Xinong Road, Yangling, Shaanxi 712100, PR China. E-mail: lydiayliu@yahoo.com
First published on 17th April 2015
Abstract
A novel electrochemically active strain D-8 was successfully isolated from rice paddy soil by combining primary enrichment on electrode, secondary biofilm selection on electrode via electrochemical consecutive selection, Fe(III)-oxide reduction, fumarate selection and a dilution–extinction plating technique. The maximum current density of 480 ± 5.5 μA cm−2 (n = 3) produced on primary biofilm increased to 1010 ± 0.4 μA cm−2 (n = 2) μA cm−2 after selection using Fe(III)-oxide, which showed that the paddy soil contained Fe(III)-reducing bacteria with a highly efficient electricity generation capability. In addition, phylogenetic analyses using the 16S rRNA gene sequence demonstrated that strain D-8 was most closely related to G. sulfurreducens PCA, with 99.53% sequence similarity. Meanwhile BOX-PCR, a rep-PCR technique, was used and provided obviously different genomic fingerprints between G. sulfurreducens and strain D-8 in some specific DNA fragments. Furthermore, strain D-8 also showed different physiological and biochemical characteristics from Geobacter sulfurreducens PCA. For example, strain D-8 is a facultative bacterium, and can use more carbon sources such as ethanol, glucose and sucrose than G. sulfurreducens PCA. These results implied that the two strains possessed distinguishable physiological, biochemical and nucleic acid characteristics. Especially, a strain D-8 biofilm on an electrode surface produced a 1.5 times higher current density of 1088 ± 7.7 μA cm−2 (n = 6) than G. sulfurreducens PCA (722 ± 2.6 μA cm−2, n = 4). Therefore, strain D-8 will be a promising bioanodic organism in microbial fuel cells.
Introduction
Microbial fuel cells (MFCs) as a novel green energy technology have a promising future due to their diverse potential applications in bioelectricity, bioremediation, biosensing, biofuel production, wastewater treatment and other new fields of application, such as desalination and the immobilization and reduction of CO2.1–7 The electrochemically catalytic activity of exoelectrogenic bacteria is the crucial factor in improving MFCs performance. Although enriched mixed-cultures show comparable or higher current-generation ability than pure cultures,8–10 single strains as model organisms can provide more clear and convenient pathways to give insights into the electron transfer mechanism between bacteria and electrodes by allowing physiological and biochemical characterization in more details than mixed-cultures. At present, well-studied pure cultures with good electricity production ability mainly include Shewanella putrefaciens or oneidensis,11,12 Geobacter sulfurreducens or metallireducens,13,14 Rhodoferax ferrireducens,15 Rhodopseudomonas palustris DX-1,16 Geothrix fermantans,17 Acidiphilium cryptum,18 and Clostridium thermocellum.19 Different species show different electricity production abilities and characteristics in MFCs. It is necessary to find more pure cultures with a highly efficient electricity generation ability. The most electricity-generating bacteria belong to the dissimilatory metal-reducing bacteria (DMRB) and they can respire with ferric iron, Mn4+, humic acids, sulfate, or a solid electrode, as the electron acceptors for the anaerobic decomposition of organic matter. These bacteria have gained significant attention and interest for applications in energy, the environment and ecology. For example, G. sulfurreducens, a strict anaerobic and iron-reducing bacterium isolated from a freshwater environment, has been adopted as the most studied culture in MFCs.3,20 G. sulfurreducens can use an anode electrode as the sole electron acceptor to completely oxidize acetate to carbon dioxide.3 The complete genome sequence of G. sulfurreducens is available, and some insights into its physiological properties during electricity production have been revealed by using genetic manipulation or linking spectral and electrochemical analysis.21,22 It was found that the multiple membrane-bound c-type cytochromes or pili greatly facilitate the electron transfer over long distances between biofilm and the electrode.21–25There exist plenty of micro-organism species in natural environments, where the pure exoelectrogenic bacteria can be obtained through a series of processes of enrichment, selection and isolation. The traditional process from dilution–extinction to testing the electricity-generation ability of strains includes picking up single colonies, transferring into growth medium and repeating the picking up and transferring until a pure culture with the ability to produce electricity is obtained, which is the most used reliable method. Meanwhile, the serial dilution process combined with a selection process using Fe(III)-oxide already shows great advantages in accelerating the isolation of exoelectrogenic bacteria. Most recently, various techniques have emerged to enhance the isolation efficiency. Particularly, the dilution–extinction U-tube method was successfully realized to isolate single exoelectrogenic bacteria.26 Another technique of biological laser printing to print living cells for isolating micro-organisms from energy-relevant environmental samples was also adopted.27 The species of electrochemically active bacteria (EAB) identified by these methods include the genus Geobacter,3,28 the genus Shewanella,29 Rhodoferax ferrireducens,15 Rhodopseudomonas palustris,16 sulfate-reducing bacteria,30 etc. Most EAB showed a good Fe(III) reduction ability.
On the other hand, wetland rice paddy fields represent about 10% of the global area of limnic ecosystems. When a paddy field is flooded and O2 is removed, the soil immediately below the surface becomes anaerobic and a community of anaerobic microbiota (comprised mainly of sulfate-reducing bacteria, iron-reducing bacteria, fermenting bacteria and methanogenic archaea) is established. The dominant species are different under different conditions. For example, Fe(III)-reducing bacteria could inhibit methane production as they compete for substrates with methane-producing bacteria, which is beneficial to rice growth. The availability of Fe(III) for microbial reduction is an important factor controlling the extent of organic matter decomposition, with Fe(III) serving as the terminal electron acceptor. Fe(III)-reducing organisms can metabolize organic matter that otherwise would be metabolized by methanogenic or sulfate-reducing food chains in the sediment. Fe(III)-reducing organisms must be capable of competing with other metabolic types of organisms for the organic matter in sediments. However, the electron transfer to oxygen will compete with electron transport to Fe(III) since aeration strongly inhibits Fe(III) reduction. Therefore, it is important to study the Fe(III)-reducing bacteria in sediment or bulk soil. G. sulfurreducens belonging to the δ-proteobacteria, gram-negative, and originally isolated from sediments at a hydrocarbon-contaminated site was studied as a typical anaerobic exoelectrogenic bacterium in MFCs.31,32 In addition, G. sulfurreducens subsp. ethanolicus was also found in lotus field mud.33 Electricity production using a MFC system based on anodes set in the rice rhizosphere and cathodes in the flooded water above the rhizosphere has already been established.34 It was found that acetate, as one of the major organic compounds in root exudates, can improve the electricity generation capability and the rice paddy soil based sediment MFC type shows promising applications. However, there are few studies on EAB based on rice paddy soil culture.34 To the best of our knowledge, the isolation of Fe(III)-oxide reducing exoelectrogenic bacteria from rice paddy field soil has not been reported up to now.
In this study, the isolation of iron reducing exoelectrogenic bacteria from paddy soil was investigated to accelerate the development of rice paddy soil based MFCs. On the other hand, it is well-known that the current density of wild type G. sulfurreducens PCA was reported to be the highest in pure cultures.35 Here a strain D-8 belonging to genus Geobacter with higher electricity-producing ability than G. sulfurreducens PCA was successfully isolated from rice paddy soil through combining several conventional isolation methods (electrode primary selection, secondary biofilm formation, Fe(III)-reduction selection and dilution–extinction procedure). Strain D-8 exhibited 99.53% similarity with G. sulfurreducens PCA using 16S rRNA gene sequence, but their BOX-PCR results showed significantly different genomic fingerprints.
Materials and methods
Electrode materials and chemicals
Polycrystalline carbon sheets (geometric surface area 1.50 cm2) were cleaned without polishing. Platinum wire with 0.3 mm diameter was used to connect the carbon materials. All chemicals were of analytical or biochemical grade.Electrochemical experiments
All electrochemical experiments were carried out on a 16-channel potentiostat in a conventional half-cell system with a three-electrode arrangement, consisting of carbon-based working electrodes, a counter electrode, and a saturated calomel electrode (SCE, Hg/Hg2Cl2 saturated KCl, 0.244 V vs. hydrogen standard electrode (SHE)) as a reference electrode. Here, all potentials refer to SCE unless otherwise stated. All chronoamperometric measurements were anaerobically conducted in a bioreactor with 30 mL solution under constant potential of 0.3 V at a temperature of 30 ± 1 °C and all current density values were normalized on the geometric surface area.36Medium, inoculum source, enrichment and biofilm formation
The soil was sampled from a rice paddy field in Shaanxi, China. For formation of primary biofilm, 5.0 g wet soil (removing rice roots and particles) was inoculated into the sealed electrochemical cell with 30 mL of nutrient medium solution containing sodium acetate saturated with nitrogen as described previously.37 The synthetic medium was prepared as in the previous report,38 containing sodium acetate (20 mM, unless otherwise stated) and a nutrient buffer solution at pH 7.2 containing 1.5 g L−1 NH4Cl, 0.3 g L−1 KH2PO4·H2O, 0.6 g L−1 KCl, 0.1 g L−1 MgCl2, 0.1 g L−1 CaCl2, and trace element (10 mL) and vitamin (10 mL) solutions sparged with highly pure nitrogen gas. All biofilm formation in this study was conducted under a potential of 0.3 V on a working electrode operated in a three-electrode system bioreactor to keep highly controlled, identical conditions unless otherwise stated, instead of a true MFC system, to avoid influence from the cathode.36 Substrate and medium replenishment was done regularly until a stable primary biofilm was formed without additional inoculation.Unless otherwise stated, operating conditions for all bacterial growth were at 30 ± 1 °C using pH 7.2 medium and all tests were conducted under sterilized conditions at least in triplicate.
The consecutive electrochemical selection procedure
After it formed, the primary biofilm was used as inoculum for the formation of all secondary biofilms.8,39 Briefly, to develop secondary biofilms, primary biofilm modified electrodes were interconnected, together with one or several blank carbon electrodes as one working electrode, and immersed into stirred, anaerobic, sterile substrate solution at a constant potential of 0.3 V.Isolation of Fe(III)-reducing exoelectrogenic culture
The rice paddy field soil was used as the original inoculum source to grow a biofilm for the exoelectrogenic bacteria enrichment. The dominant exoelectrogenic bacteria of the as-formed biofilm was selected through the consecutive electrochemical selection procedure for growing secondary biofilms and then was further selected using amorphous ferric oxyhydroxide (FeOOH) as the sole electron acceptor. In detail, the suspension medium consisting of secondary biofilm was transferred into red colored solutions in bottles containing Fe(III)-oxide and incubated at 30 °C.When the medium color changed from red to black, the black medium was used as inoculum again for iron-reducing bacteria growth for several generations. Then the growth well medium was used for ten times serial dilutions up to 108 times. Each diluted solution was plated on a Petri dish with solid media (1.5% agar) for isolation. All experiments were carried out under anaerobic conditions and the inoculated plates were incubated at 30 ± 1 °C. After the colonies could be observed by eye, single colonies were picked up using a sterile pipette tip and transferred into liquid growth medium containing the same constituents as the solid medium except agar and incubated at 30 ± 1 °C for the next steps of testing the electricity-producing capability and identifying single strains. The whole isolation procedure was recycled until all the single colonies were purified to show a similar appearance and could produce electricity. Meanwhile, the isolated colonies were also picked up and restreaked onto agar plates for further confirming the purified bacterium.
G. sulfurreducens and biofilm formation
G. sulfurreducens PCA, purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, was cultured in growth medium containing 1.64 g L−1 sodium acetate, 1.5 g L−1 NH4Cl, 0.3 g L−1 KH2PO4·H2O, 0.6 g L−1 KCl, 0.1 g L−1 MgCl2, 0.1 g L−1 CaCl2 and 4.64 g L−1 fumaric acid, and 10 mL of trace element solution and 10 mL of vitamin solution were added as previously described.40 The medium was adjusted to pH 7.2 and dissolved oxygen was removed before use. 10% culture was inoculated into a three-electrode electrochemical cell and 0.3 V potential was applied to grow the G. sulfurreducens biofilm.36 All tests were performed at least in triplicate. For further confirming the strain of G. sulfurreducens which was used in this study, its gene sequence was analyzed and it was found that its 16S rRNA gene sequence is completely related to G. sulfurreducens PCA (100% sequence similarity, data not shown).16S rRNA gene sequencing and DGGE analysis
For identification of the isolated strain, the genomic DNA of each fumarate-acetate-growth strain was extracted using a BacteraGen DNA Kit (CWBIO, China) according to manufacturer’s instructions. The amplification of 16S rRNA genes was performed using universal primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′). The PCR amplification was performed in a 20 μL reaction vessel containing 1 μL of DNA, 1 μL of each primer, 10 μL premix (2 × Taq PCR Mix) and 7 μL ddH2O. PCR amplification was carried out on an XP Cycler gene amplification instrument (BIOER, China) with an initial denaturation of DNA for 5 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 1 min at 55 °C and 1 min at 72 °C, and then final extension for 10 min at 72 °C. The PCR products were purified using an E.Z.N.A.® Gel extraction kit (OMEGA) and then were examined using agarose gel electrophoresis prior to analyzing the sequence.The 16S rRNA gene sequence obtained was compared to the sequences of the most closely related strains in the GenBank database by using the BLAST program. Then the retrieved sequences were aligned using the Clustal X 1.83 program. A neighbor-joining phylogenetic tree was constructed using the Molecular Evolutionary Genetics Analysis package (MEGA, version 5).41 A bootstrap analysis was based on 1000 replicates. For denaturing gradient gel electrophoresis (DGGE) analysis, the extracted DNA was first amplified using the universal primers 27F and 1492R, and then the products were amplified again with the primer set 357F, containing a GC clamp (5′-CGC CCG CCG CGC GGC GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG-3′), and 518R (5′-ATT ACC GCG GCT GCT GG-3′). The PCR amplifications were performed as described above. Then the PCR products (15 μL) were separated using 8% (wt/vol) polyacrylamide gels with a denaturant gradient between 40% and 60%. The Dcode Universal Mutation Detection System (Bio-Rad, CA, USA) was used for DGGE, which was first run in 1 × Tris-acetate-EDTA buffer at 50 V for 30 min and subsequently at 80 V for 12 h (60 °C). After electrophoresis, the gels were stained using ethidium bromide (EB) for 15 min before the DNA bands were observed with a Gel-Doc image analyzer (Bio-Rad Laboratories).
BOX-PCR analysis and strain identification
The DNA extracted from the strain D-8 bacteria was used for PCR amplification by using the BOXA1R primer: (5′-CTA CGG CAA GGC GAC GCT GAC G-3′).42 Amplification reactions were performed in volumes of 20 μL, containing 2 μL of a single BOXA1R primer, 0.5 μL of dNTPs, 2 μL 10 × PCR reaction buffer, 0.2 μL Taq DNA polymerase, 1 μL of chromosomal DNA and 14.3 μL deionized water. PCR amplifications were performed with a DNA thermal cycler (BIOER, China) programmed for an initial denaturation step of 7 min at 95 °C, followed by 32 cycles of 1 min at 94 °C, 1 min at 53 °C, 8 min at 65 °C, with a final elongation of 15 min at 65 °C. After the reactions, 10 μL of the BOX-PCR products were separated by gel electrophoresis in a 1.5% agarose gel for 4 h at a constant voltage of 4 V cm−1 in 1 × TAE. The BOX-PCR products were photographed by using a Gel imaging system (Bio-Rad, Laboratories) after staining with ethidium bromide under ultraviolet light. Negative controls were included in each experiment, replacing the chromosomal DNA with deionized water. The BOX-PCR analysis was performed in at least three replicates in separate experiments to confirm the reliability of the results.Growth using alternative electron donors
The medium containing 1.64 g L−1 sodium acetate, 1.5 g L−1 NH4Cl, 0.3 g L−1 KH2PO4·H2O, 0.6 g L−1 KCl, 0.1 g L−1 MgCl2, 0.1 g L−1 CaCl2 and 4.64 g L−1 fumaric acid, and 10 mL of trace element solution and 10 mL of vitamin solution as previously described40 was used to grow strain D-8 and Geobacter sulfurreducens PCA under identical conditions. When the respective OD of strain D-8 and G. sulfurreducens PCA was around 0.4 and 0.6, the two cultures were centrifugated and the supernatant was poured out. After the residue was rinsed 5 times using the above medium without acetate to remove acetate completely, the above medium with the respective electron donor instead of acetate was used for each incubation. When the OD value arrived at a stable point, 2% inoculation was conducted again into the medium with the same substrate for recording OD value change with time. Control experiments were simultaneously conducted to avoid possible contamination during operation. All the operations were done under sterilized conditions by autoclaving or membrane filtration with a pore diameter of 0.22 μm.The assay of acid production
The medium consisted of 0.2% peptone, 1% carbohydrate and 0.5% NaCl. It was adjusted to pH 6.8–7.0 and 0.3% bromothymol blue sodium salt (BTB) solution (dissolved in 1% ethanol) was added. 2% inoculation was used and the medium was incubated at 30 °C.Influence of pH, temperature and salinity on the growth of G. sulfurreducens and strain D-8
The growth curves were measured by OD value changes of G. sulfurreducens or strain D-8 incubated in medium (pH 7.2) containing 20 mM acetate as an electron donor at regular intervals at 30 °C. The influence of various pH values was tested using OD values of G. sulfurreducens or strain D-8 in medium containing 20 mM acetate as an electron donor during incubation at the corresponding pH for 48 h at 30 °C. Influence from various temperatures was studied using OD values of G. sulfurreducens or strain D-8 in medium (pH 7.2) containing 20 mM acetate as an electron donor during incubation at the corresponding temperature for 48 h. The growth curves with different salinity concentrations were obtained from OD value changes of G. sulfurreducens or strain D-8 in medium (pH 7.2) containing 20 mM acetate as an electron donor and different NaCl concentrations at regular intervals at 30 °C. All OD measurements were conducted using 2% inoculation and monitored at 600 nm in triplicate.Morphological analysis and sample preparation
After the biofilm attached electrode was washed three times, it was fixed in 2.5% glutaraldehyde in phosphate buffer (0.1 M, pH 7.2) for 12 h at 4 °C and was immersed in buffer supplemented with 1% OsO4 for 30 min. Then a graded series of aqueous ethanol solutions (10–100%) and a graded series of tertiary butanol (25–100%) were successively used to dehydrate samples. Finally samples were freeze dried via tertiary butanol for 2.5 h and sputter coated with gold prior to scanning electron microscopy (SEM) observation. The examination was done in a Hitachi S-3400N SEM (Hitachi, Tokyo, Japan) at 15 kV.A Nikon A1 spectral imaging confocal microscope (Nikon, Japan) was used to image biofilm-covered electrodes. When the biofilm produced maximum current, the biofilm-covered electrodes were gently washed in growth medium to remove planktonic cells and then stained with the Hoechst 33342 (fluorescent DNA dye),43 incubated for 15 min in the dark, rinsed with growth medium, and placed on a microscope slide. One laser wavelength, 405 nm, was used to excite the stains.
Results and discussion
Primary enrichment of exoelectrogenic bacteria from paddy soil and secondary biofilm formation using electrochemical selection procedure
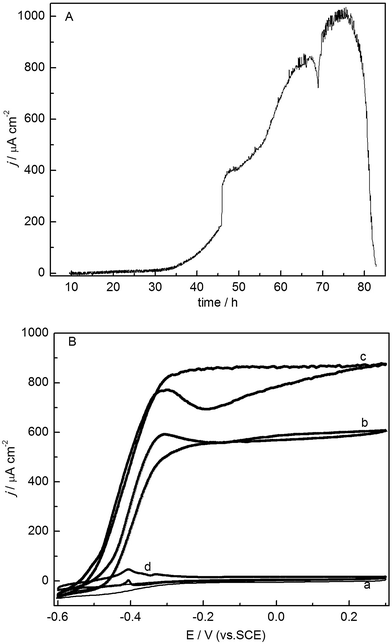
The primary electrochemically active biofilm accumulation and enrichment were studied on a carbon sheet electrode using chronoamperometry under constant potential. Fig. 1A shows that after inoculation the current density stayed around 0 μA for about 24 h and then it rapidly increased to 80 μA cm−2 at 60 h. This result implied that the primary biofilm could form on the electrode surface at 60 h. In order to further confirm it, at 63 h the used medium was replaced completely using fresh medium containing acetate without additional inoculation. Meanwhile, the planktonic cells were also removed. As shown in Fig. 1A, the current in the medium without planktonic cells increased to 120 μA cm−2 at 90 h, which confirmed that the produced current resulted from the attached biofilm on the electrode. Other medium exchanges were successively conducted to provide enough nutrients and acetate (as indicated by arrows at 113 h, 160 h and 266 h, respectively) to further accelerate the biofilm growth. As expected, the catalytic current significantly increased from the original 175 μA cm−2 to 480 ± 5.5 μA cm−2 (n = 3) with regular medium exchanges. After the fourth exchange, the current density from the primary biofilm showed no obvious improvement, which might be due to the biofilm growth over a long time resulting in a low bacterial activity. The cyclic voltammograms of biofilms at various growth stages were also measured to evaluate the performance of the electrochemically active biofilms (Fig. 1B). Before inoculation there were no obvious redox peaks observed (curve a in Fig. 1B). At 63 h, the voltammogram showed a pair of sluggish redox peaks (curve b in Fig. 1B), exhibiting higher charging current than that at the inoculation point. These data also indicated that the primary EAB had already formed on the electrode surface after inoculation for 63 h as the higher changing current might result from the attached cells on the electrode surface. Here the formed biofilm at this stage is denominated as primary soil-culture. With regular medium exchanges the voltammograms showed an increased catalytic behavior (curves c and d in Fig. 1B).A consecutive electrochemical selection procedure was adopted to improve the electrochemical activity8,37 in this study. From Fig. 1A, the maximum current density on a series of blank electrodes interconnected with the primary biofilm covered electrode was considerably increased to 676 ± 3.1 μA cm−2 (n = 5) after one month (the formed secondary biofilm at this stage is denominated as secondary soil-culture). It can be seen that the electricity-production capability of secondary biofilms based on rice paddy soil was further improved by using an electrochemical selection procedure because the non-electrochemically active cells can be rejected during this process.37 Especially, the catalytic currents on the voltammograms on secondary biofilms were greatly enhanced and the voltammogram shape was also changed to a typical sigmoidal shape (curve e in Fig. 1B). These are due to the effective selection of exoelectrogenic cells and the elimination of those non-electrochemically active cells possibly entrapped in the biofilm during the consecutive electrochemical procedure.37 After substrate exhaustion, the voltammogram from the secondary soil-culture biofilm showed two pairs of obvious redox peaks with formal potential of −0.42 and −0.46 V (curve f in Fig. 1B), which did not occur before inoculation (curve a in Fig. 1B). In addition, the electrochemical characterization of soil-culture biofilms under substrate exhaustion is similar with that on the typical exoelectrogenic pure culture of wild-type G. sulfurreducens PCA.44 Fig. 1A and B revealed that a mixed-culture biofilm based on rice paddy soil already formed on the electrode surface and possessed a catalytic ability up to 676 ± 3.1 μA cm−2 (n = 5) in secondary biofilms.
The biocatalytic performance of the soil-culture biofilm after selection using ferric oxyhydroxide (Fe-soil-culture)
Iron geochemistry is an important aspect in anoxic subsurface environments. Rice paddy field soil already exhibits plenty of iron reducing bacteria.45 In addition, the ferric oxyhydroxide selection process for selecting and isolating EAB demonstrated good reliability.46 Here, the secondary soil-culture was further selected using ferric oxyhydroxide as an electron acceptor. When the secondary biofilm attached on the electrode was inoculated into red Fe(III)-oxide medium, and incubated for one week, the formation of black precipitates from initially red colored solutions was observed. No black precipitation was observed in the control experiment without the biofilm related culture inoculation. The black precipitates, mainly magnetite (Fe3O4) and siderite (FeCO3) could indicate the Fe(III) reduction by the inoculated organisms from the biofilm in the solution.47 The resulting culture was denominated as Fe-soil-culture in this study. Fig. 2A shows the chronoamperometric curve of the secondary soil-culture after selection using ferric oxyhydroxide. The similar sigmoidal shape of the voltammogram in the presence of acetate was observed but with a maximum catalytic current density up to 1010 ± 0.4 μA cm−2 (n = 2), which was much higher than the 676 ± 3.1 μA cm−2 (n = 5) before Fe(III)-oxide reduction selection. There are two similar pairs of obvious redox peaks under acetate exhaustion with that before Fe(III)-oxide reduction selection (Fig. 2B). It can be seen that the catalytic current increased by 42.8% on Fe-soil-culture biofilm after selection, might be due to the ferric oxyhydroxide as an electron acceptor can further select the exoelectrogenic bacteria with the ability to reduce ferric oxyhydroxide and meanwhile further eliminate those non-exoelectrogenic cells randomly entrapped in the biofilm. In addition, stirring caused the increase of catalytic current by 40% (curve c in Fig. 2B) in comparison with that in quiescent solution.8 It can be found that the biocatalytic performance of the mixed biofilm based on rice paddy soil was increased nearly to 2 times higher than that of primary biofilm after adopting the electrochemical consecutive selection and ferric oxyhydroxide selection procedures.Isolation of a single culture from Fe-soil-culture and strain analysis
The Fe-soil-culture was transferred into fumarate medium and was isolated with agar media on a Petri dish using dilution–extinction method. After the colonies could be observed by eye, single colonies were picked up using a sterile pipette tip and transferred into liquid growth medium containing the same constituents as the solid medium except agar and incubated at 30 ± 1 °C. The whole isolation procedure of recycling steps between picking up single colonies and transferring into liquid growth medium was repeated until single colonies were measured to show completely identical 16S rRNA sequences. Meanwhile, the Streak-Plate technique was also adopted and further confirmed the purification of the isolated strain. Here three separate single colonies named D-8, D-10 and D-12 as the models of isolated strains were showed for their electricity production abilities.The PCR of the 16S rRNA gene from each colony showed the same single band (Fig. S1†). Through 16S rRNA gene sequence analysis, a completely identical gene sequence of strains D-8, D-10 and D-12 was obtained. Here strain D-8 as a model of the isolated strains was further studied.
The nearly complete 16S rRNA gene sequence (at least 1400 bases sequenced) was amplified by PCR using strain D-8 genome DNA as a template and 16S rRNA universal primers of 27F/1492R. The sequence similarity of the 16S rRNA gene was compared with those reference organisms obtained from GenBank database and EzBioCloud database. Geobacter sulfurreducens PCA (AE017180) was its nearest neighbor, with a sequence similarity of 99.53%. A constructed phylogenetic tree as shown in Fig. 3 showed the relationship between strain D-8 (D-10, or D-12) and other related species, indicating that these organisms belong to the genus Geobacter (Fig. 3). The completely identical gene sequence of the three strains (D-8, D-10 and D-12) implied that the isolated strain could be the most dominant exoelectrogenic species with iron reducing ability in the enriched rice paddy soil community. The low taxon richness in the biofilm community after a month of selection using a potentiostat was already reported.48 These results are consistent with the report that iron reducing organisms are abundant in rice paddy soil and play an important role in the rice paddy environment and iron cycling.
Changes in the microbial community of biofilms during the enrichment and selection periods were analyzed by DGGE profiles. Based on the migration distance, intensities, and similarities between the lanes on the DGGE gel, the banding patterns of the biofilm showed great differences before and after Fe(III)-oxide selection (Fig. S2†). From DGGE analysis, two strong bands (1 and 2) and several faint bands (3 and 4) were observed before selection using Fe(III)-oxide (Lane C in Fig. S2†), while after Fe(III)-oxide selection, one of the faint bands, 4, becomes the most prominent band (Lane B). This implied that the Fe(III)-oxide reducing bacteria became the dominant species after selection using Fe(III)-oxide reduction by eliminating the non-Fe(III) reducing bacteria. From the higher current of the Fe-soil-culture biofilm than that of the soil-culture biofilm in Fig. 2, the non-Fe(III) reducing bacteria, being the majority of the population before Fe(III)-oxide reducing selection, could have lower electricity production capability than Fe(III) reducing bacteria, which resulted in the low current. As expected, one single bright band 4 was observed for strain D-8 (Lane A), showing that strain D-8 was the dominant Fe(III)-oxide reducing bacterium in the Fe-soil-culture and Fe(III)-oxide reducing selection is an effective selection process for isolating Fe(III)-oxide reducing strains. Although a single band does not always represent a single bacterial strain in DGGE analysis due to possible mis-incorporation or mis-reading during PCR and sequencing steps,49 this band was excised and sequenced, and showed 100% similarity of 16S rRNA gene fragments with strain D-8. The isolation and identification of other non-Fe(III)-oxide reducing electricity producing bacteria involved in the soil-culture is required in a further study.
BOX-PCR analysis
Besides 16S rRNA analysis, rep-PCR based on repetitive sequence as a rapid and highly discriminatory screening technique such as ERIC-, BOX-, and REP-PCR, can be used to distinguish bacteria at the species, (sub)species and strain level.50,51 Among them REP-PCR and BOX-PCR have already been used to identify and characterize some Geobacter isolates and thermophilic bacteria.33,52,53 In our study, there was strong similarity (more than 99%) between the strain D-8 and G. sulfurreducens PCA based on the 16S rRNA gene (Fig. 3). Due to the polymorphism of the 16S rRNA gene it was considered insufficient to discriminate and identify some closely related strains.54 Here in order to further distinguish strain D-8 and G. sulfurreducens, BOX-PCR was conducted using the BOXA1R primer. From Fig. 4, approximately 8–12 fragments of the size 0.5–5 kb were observed using BOX-PCR, while the two strains had distinctly different patterns. This indicated that even though the strain D-8 has more than 99% similarity with the described G. sulfurreducens based on the 16S rRNA gene, rep-PCR band patterns for both strains revealed distinguishable genomic DNA fingerprints. Reproducible results were obtained from the completely identical band patterns in triplicate. These data show that there is significant difference between G. sulfurreducens PCA and strain D-8 in some specific DNA fragments. | ||
| Fig. 4 BOX-PCR fingerprints: Lane N, negative control; Lane 1, G. sulfurreducens PCA; Lane 2, strain D-8, Lane M, DNA marker DS5000. | ||
Growth analysis using various electron donors for strain D-8 and G. sulfurreducens PCA
Various electron donors were used for both G. sulfurreducens and strain D-8 growth using fumarate as an electron acceptor. The OD at 600 nm was monitored at regular intervals and is shown in Fig. 5. G. sulfurreducens can grow very well using acetate and pyruvate, while it cannot use formate, ethanol, glycerol, glucose, sucrose or starch (Fig. 5A). By contrast, glucose and sucrose can be used as electron donors by strain D-8 but not G. sulfurreducens in simultaneous experiments under identical conditions (Fig. 5B). Meanwhile, the changes of the OD values were neglectable in the control experiments without inoculation, which also eliminated the possibility of contamination during operating steps. It can be seen that strain D-8 can utilize more substrates including acetate, pyruvate, formate, ethanol, glycerol, glucose, sucrose and starch than G. sulfurreducens.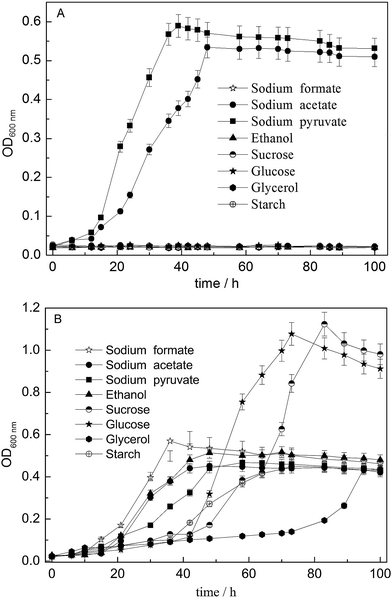 | ||
| Fig. 5 The growth curves of G. sulfurreducens (A) and strain D-8 (B) utilizing various electron donors. Error bars are 5%. | ||
Physiological and biochemical features of strain D-8 in comparison to G. sulfurreducens PCA
The growth at different temperatures, pH values, salt concentrations or in the presence of various substrates was investigated for G. sulfurreducens and strain D-8 under identical conditions. From Fig. 6A, both G. sulfurreducens and strain D-8 entered stationary phase at ca. 30 h with respective maximum OD values of 0.58 and 0.42. The optimum pH of G. sulfurreducens and strain D-8 was pH 7.2 and 7.5, respectively. The optimum temperature for G. sulfurreducens is 40 °C and for strain D-8 is around 35 °C (Fig. 6C). Fig. 6D showed that G. sulfurreducens can grow in medium with 1% NaCl, which is consistent with the report of G. sulfurreducens tolerating about 1.5% NaCl.32 By comparison, strain D-8 cannot grow even with 1% NaCl. These results indicated that the characteristics of G. sulfurreducens PCA and strain D-8 at optimum pH, temperature and salt tolerance are different from each other.BTB was used as indicator to assay acid production in the medium as shown in Fig. S3.† The strain D-8 medium became a yellowish-green color after 24 h and bright yellow after 72 h. Meanwhile, G. sulfurreducens retained a greenish sheen even after 72 h. This meant that strain D-8 produced an acid reaction in the medium and had the ability of glycolysis, while G. sulfurreducens did not.
Using the Gram-staining method, strain D-8 is Gram-negative just like G. sulfurreducens PCA (data not shown).
The amylohydrolysis and oxygen tolerance were analyzed using iodine as an indicator as shown in Fig. S4.† The tube containing strain D-8 became colorless, but the G. sulfurreducens tube remained blue. This indicated that strain D-8 can hydrolyze starch, while G. sulfurreducens cannot. On the other hand, in anaerobic conditions, both G. sulfurreducens and strain D-8 can reduce red ferric oxyhydroxide (FeOOH) to produce a black suspension. Fig. S4† shows strain D-8 can reduce amorphous ferric oxyhydroxide (FeOOH) after its medium was saturated by air filtered using a membrane with pore diameter of 0.22 μm for up to 10 min. By contrast, the bottle containing medium with G. sulfurreducens pumped with air even for 5 min did not show any obvious iron reduction phenomena, which is consistent with the strict anaerobic characteristics of G. sulfurreducens PCA. Especially, strain D-8 can be plated on a Petri dish with solid media to grow into colonies in the presence of air, where the colonies appeared wrinkled and flattened, showing distinctly different characteristics from the colonies being smooth and convex under anaerobic conditions (data not shown). The strictly anaerobic G. sulfurreducens PCA cannot grow in the presence of air. From these results, the strain D-8 is a facultative culture rather than the strictly anaerobic.
Electricity-generation capability of strain D-8
The three separated single colonies of D-8, D-10 and D-12 were randomly picked for simultaneous gene sequencing and electricity production ability testing. The three single strains with identical gene sequence also showed a similar maximum catalytic current density (Fig. 7 and Table 1). The electricity producing abilities of strains D-8, D-10, D-12 and their closest species of G. sulfurreducens PCA were compared under identical conditions (in medium with pH 7.2 and at 30 °C) as shown in Fig. 7. Table 1 lists the maximum current densities achieved from a number of independent chronoamperometric curves of strains of D-8, D-10, D-12 and G. sulfurreducens. Unexpectedly, the average maximum current density of 1088 ± 7.7 μA cm−2 (n = 6) of strain D-8 was 1.5 times higher than the 722 ± 2.6 μA cm−2 (n = 4) of the G. sulfurreducens biofilm. On the other hand, the isolated strain D-8 exhibited comparable catalytic current ability with the Fe-soil-culture (1010 ± 0.4 μA cm−2, n = 2) before isolation. It can be seen that strain D-8 could be the dominant strain in the Fe-soil-culture. Fig. 7A shows the typical chronoamperometric curves from single strain D-8 and G. sulfurreducens biofilms with each in duplicate. Accordingly, the voltammograms of strain D-8 and G. sulfurreducens biofilms were also compared. From their chronoamperometric curves and voltammograms (Fig. 7A–C), strain D-8 biofilm produced more than 1.5 times higher catalytic current density than that of the G. sulfurreducens biofilm. However, under non-catalytic conditions, their voltammograms showed two pairs of obvious redox peaks at similar potential positions (curve d in the insets of Fig. 7B and C). In addition, the electricity generation of strain D-8 utilizing ethanol, pyruvate, glycerol and glucose was also measured using chronoamperometry as shown in Fig. 7D (each substrate was done in duplicate). It can be seen that pyruvate has a little lower electricity generation ability (940 μA cm−2) than acetate or ethanol, while using glucose and glycerol gave lower currents of around, 390 and 300 μA cm−2, respectively. Although the current densities of strain D-8 from glucose and glycerol were much lower than acetate or ethanol, they implied that strain D-8 can utilize glucose and glycerol to produce electricity, which is consistent with the growth curves in Fig. 4B. By comparison, G. sulfurreducens cannot utilize substrates such as ethanol, glucose and glycerol. These results further indicated that strain D-8 can utilize more carbon sources to generate electricity than G. sulfurreducens PCA.| No. | Current density/μA cm−2 | ||||
|---|---|---|---|---|---|
| Before isolation | After isolation | G. sulfurreducens | |||
| Primary soil-culture | Secondary soil-culture | Fe-soil-culture | Strain D-8 (D-10, D-12) | ||
| a Notes: “—” represents no data shown. | |||||
| 1 | 450 | 656 | 1007 | 1002 | 722 |
| 2 | 500 | 662 | 1013 | 1109 | 705 |
| 3 | 490 | 674 | — | 1176 | 711 |
| 4 | — | 703 | — | 1192 | 748 |
| 5 | — | 687 | — | 1010 | — |
| 6 | 1040 | — | |||
| Average value | 480 | 676 | 1010 | 1088 | 722 |
| RSD% | 5.5 | 3.1 | 0.4 | 7.7 | 2.6 |
Morphological analysis of biofilms
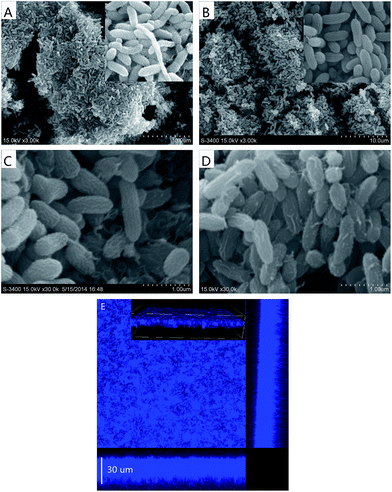
The morphologcial characteristics of soil-culture biofilms before and after isolation are shown in Fig. 8. It can be clearly seen that there is a huge biofilm covering the entire electrode surface in both cultures after growth for 53 h. The soil-culture biofilm exhibited diverse sizes and shapes (Fig. 8A), and about 1.5 μm long rod shaped bacteria were dominant. By contrast, the biofilm of strain D-8 presented in uniform sizes and shapes after isolation (Fig. 8B and C). In addition, a comparison between strain D-8 and G. sulfurreducens PCA growth medium was also conducted under identical conditions (Fig. 8C and D). Strain D-8 appeared rod shaped, about 1.5 μm in length and 0.5 μm in diameter, which is similar with G. sulfurreducens PCA (Fig. 8D). The confocal image produced by staining with Hoechst 33342 showed that the thickness of mature strain D-8 biofilm is around 30 μm (Fig. 8E), which also is similar with G. sulfurreducens PCA (data not shown).Conclusion
In this study, strain D-8 with a high electricity generation capability was successfully isolated from rice paddy soil. Through phylogenetic analyses using the 16S rRNA gene sequence, strain D-8 demonstrated 99.53% similarity to G. sulfurreducens PCA belonging to the genus Geobacter, while their BOX-PCR genomic fingerprints are distinguishable. On the other hand, strain D-8 demonstrated different physiological and biochemical characteristics from G. sulfurreducens PCA. Especially, strain D-8 is facultative not strictly anaerobic and can use more carbon sources including ethanol, glucose, sucrose, etc. These results implied that strain D-8 exhibited different physiological, biochemical and nucleic acid characteristics from G. sulfurreducens PCA. Most importantly, strain D-8 biofilm produced 1.5 times higher current than that of G. sulfurreducens PCA. The further identification of strain D-8 is in progress. Our study demonstrated that there are superior strains in the natural environment with desired properties for applications in MFCs.Acknowledgements
We thank Prof. Xia Huang of the Tsinghua University for kindly providing G. sulfurreducens culture. This work was financially supported by a project from the National Natural Science Foundation of China (no. 21375107).References
- U. Schröder, Phys. Chem. Chem. Phys., 2007, 9, 2619–2629 RSC.
- L. M. Tender, C. E. Reimers, H. A. Stecher, D. E. Holmes, D. R. Bond, D. A. Lowy, K. Pilobello, S. J. Fertig and D. R. Lovley, Nat. Biotechnol., 2002, 20, 821–825 CrossRef CAS PubMed.
- D. R. Bond, D. E. Holmes, L. M. Tender and D. R. Lovley, Science, 2002, 295, 483–485 CrossRef CAS PubMed.
- X. C. Abrevaya, N. J. Sacco, M. C. Bonetto, A. Hilding-Ohlsson and E. Corton, Biosens. Bioelectron., 2015, 63, 580–590 CrossRef CAS PubMed.
- M. C. Hatzell, R. D. Cusick and B. E. Logan, Energy Environ. Sci., 2014, 7, 1159–1165 CAS.
- W. W. Li, H. Q. Yu and Z. He, Energy Environ. Sci., 2014, 7, 911–924 CAS.
- H. Ren, C. I. Torres, P. Parameswaran, B. E. Rittmann and J. Chae, Biosens. Bioelectron., 2014, 61, 587–592 CrossRef CAS PubMed.
- Y. Liu, D. D. Deng and X. J. Lan, Electrochim. Acta, 2015, 155, 327–334 CrossRef CAS.
- S. I. Ishii, K. Watanabe, S. Yabuki, B. E. Logan and Y. Sekiguchi, Appl. Environ. Microbiol., 2008, 74, 7348–7355 CrossRef CAS PubMed.
- V. R. Nimje, C.-Y. Chen, H.-R. Chen, C.-C. Chen, Y. M. Huang, M.-J. Tseng, K.-C. Cheng and Y.-F. Chang, Bioresour. Technol., 2012, 104, 315–323 CrossRef CAS PubMed.
- B. R. Ringeisen, E. Henderson, P. K. Wu, J. Pietron, R. Ray, B. Little, J. C. Biffinger and J. Jones-Mehan, Environ. Sci. Technol., 2006, 40, 2629–2634 CrossRef CAS PubMed.
- H. J. Kim, H. S. Park, M. S. Hyun, I. S. Chang, M. Kim and B. H. Kim, Enzyme Microb. Technol., 2002, 30, 145–152 CrossRef CAS.
- B. K. Min, S. A. Cheng and B. E. Logan, Water Res., 2005, 39, 1675–1686 CrossRef CAS PubMed.
- D. R. Bond and D. R. Lovley, Appl. Environ. Microbiol., 2003, 69, 1548–1555 CrossRef CAS PubMed.
- S. K. Chaudhuri and D. R. Lovley, Nat. Biotechnol., 2003, 21, 1229–1232 CrossRef CAS PubMed.
- D. F. Xing, Y. Zuo, S. A. Cheng, J. M. Regan and B. E. Logan, Environ. Sci. Technol., 2008, 42, 4146–4151 CrossRef CAS PubMed.
- M. G. Mehta-Kolte and D. R. Bond, Appl. Environ. Microbiol., 2012, 78, 6987–6995 CrossRef CAS PubMed.
- A. P. Borole, H. O’Neill, C. Tsouris and S. Cesar, Biotechnol. Lett., 2008, 30, 1367–1372 CrossRef CAS PubMed.
- S. H. Shin, J. G. Zeikus and M. K. Jain, Appl. Microbiol. Biotechnol., 2002, 58, 476–481 CrossRef PubMed.
- R. Mahadevan, D. R. Bond, J. E. Butler, A. Esteve-Nuñez, M. V. Coppi, B. O. Palsson, C. H. Schilling and D. R. Lovley, Appl. Environ. Microbiol., 2006, 72, 1558–1568 CrossRef CAS PubMed.
- N. S. Malvankar, M. T. Tuominen and D. R. Lovley, Energy Environ. Sci., 2012, 5, 8651–8659 CAS.
- Y. Liu, H. Kim, R. R. Franklin and D. R. Bond, ChemPhysChem, 2011, 12, 2235–2241 CrossRef CAS PubMed.
- H. Richter, K. P. Nevin, H. F. Jia, D. A. Lowy, D. R. Lovley and L. M. Tender, Energy Environ. Sci., 2009, 2, 506–516 CAS.
- Y. Liu and D. R. Bond, ChemSusChem, 2012, 5, 1047–1053 CrossRef CAS PubMed.
- G. Reguera, K. P. Nevin, J. S. Nicoll, S. F. Covalla, T. L. Woodard and D. R. Lovley, Appl. Environ. Microbiol., 2006, 72, 7345–7348 CrossRef CAS PubMed.
- Y. Zuo, D. F. Xing, J. M. Regan and B. E. Logan, Appl. Environ. Microbiol., 2008, 74, 3130–3137 CrossRef CAS PubMed.
- B. R. Ringeisen, S. E. Lizewski, L. A. Fitzgerald, J. C. Biffinger, C. L. Knight, W. J. Crookes-Goodson and P. K. Wu, Electroanalysis, 2010, 22, 875–882 CrossRef CAS.
- J. D. Coates, E. J. P. Phillips, D. J. Lonergan, H. Jenter and D. R. Lovley, Appl. Environ. Microbiol., 1996, 62, 1531–1536 CAS.
- E. Marsili, D. B. Baron, I. D. Shikhare, D. Coursolle, J. A. Gralnick and D. R. Bond, PNAS, 2008, 105, 3968–3973 CrossRef CAS PubMed.
- D. J. Lee, X. Liu and H. L. Weng, Bioresour. Technol., 2014, 156, 14–19 CrossRef CAS PubMed.
- D. R. Lovley, Nat. Rev. Microbiol., 2003, 1, 35–44 CrossRef CAS PubMed.
- F. Caccavo, D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz and M. J. McInerney, Appl. Environ. Microbiol., 1994, 60, 3752–3759 CAS.
- S. Viulu, K. Nakamura, A. Kojima, Y. Yoshiyasu, S. Saitou and K. Takamizawa, J. Gen. Appl. Microbiol., 2013, 59, 325–334 CrossRef CAS PubMed.
- N. Kaku, N. Yonezawa, Y. Kodama and K. Watanabe, Appl. Microbiol. Biotechnol., 2008, 79, 43–49 CrossRef CAS PubMed.
- K. P. Nevin, H. Richter, S. F. Covalla, J. P. Johnson, T. L. Woodard, A. L. Orloff, H. Jia, M. Zhang and D. R. Lovley, Environ. Microbiol., 2008, 10, 2505–2514 CrossRef CAS PubMed.
- H. Yi, K. P. Nevin, B.-C. Kim, A. E. Franks, A. Klimes, L. M. Tender and D. R. Lovley, Biosens. Bioelectron., 2009, 24, 3498–3503 CrossRef CAS PubMed.
- Y. Liu, F. Harnisch, K. Fricke, R. Sietmann and U. Schroder, Biosens. Bioelectron., 2008, 24, 1006–1011 CrossRef CAS PubMed.
- J. R. Kim, B. Min and B. E. Logan, Appl. Microbiol. Biotechnol., 2005, 68, 23–30 CrossRef CAS PubMed.
- Y. Liu, F. Harnisch, K. Fricke, U. Schröder, V. Climent and J. M. Feliu, J. Biosens. Bioelectron., 2010, 25, 2167–2171 CrossRef CAS PubMed.
- D. R. Lovley, R. C. Greening and J. G. Ferry, Appl. Environ. Microbiol., 1984, 48, 81–87 CAS.
- N. Saitou and M. Nei, Mol. Biol. Evol., 1987, 4, 406–425 CAS.
- T. Koeuth, J. Versalovic and J. R. Lupski, Genome Res., 1995, 5, 408–418 CrossRef CAS PubMed.
- Z. M. Puyen, E. Villagrasa, J. Maldonado, I. Esteve and A. Sole, Curr. Microbiol., 2012, 64, 75–80 CrossRef CAS PubMed.
- Y. Liu, H. Kim, R. Franklin and D. R. Bond, Energy Environ. Sci., 2010, 3, 1782–1788 CAS.
- R. Grosskopf, P. H. Janssen and W. Liesack, Appl. Environ. Microbiol., 1998, 64, 960–969 CAS.
- A. J. Wang, D. Sun, N. Q. Ren, C. Liu, W. Z. Liu, B. E. Logan and W. M. Wu, Bioresour. Technol., 2010, 101, 5733–5735 CrossRef CAS PubMed.
- E. E. Roden and D. R. Lovley, Appl. Environ. Microbiol., 1993, 59, 734–742 CAS.
- A. S. Commault, G. Lear, M. A. Packer and R. J. Weld, Bioresour. Technol., 2013, 139, 226–234 CrossRef CAS PubMed.
- H. Sekiguchi, N. Tomioka, T. Nakahara and H. Uchiyama, Biotechnol. Lett., 2001, 23, 1205–1208 CrossRef CAS.
- J. L. W. Rademaker, B. Hoste, F. J. Louws, K. Kersters, J. Swings, L. Vauterin, P. Vauterin and F. J. de Bruijn, Int. J. Syst. Evol. Microbiol., 2000, 50, 665–677 CrossRef CAS PubMed.
- G. Nick, M. Jussila, B. Hoste, R. M. Niemi, S. Kaijalainen, P. de Lajudie, M. Gillis, F. J. de Bruijn and K. Lindstrom, Syst. Appl. Microbiol., 1999, 22, 287–299 CrossRef.
- A. Adiguzel, H. Ozkan, O. Baris, K. Inan, M. Gulluce and F. Sahin, J. Microbiol. Methods, 2009, 79, 321–328 CrossRef CAS PubMed.
- Y. Sung, K. F. Fletcher, K. M. Ritalaliti, R. P. Apkarian, N. Ramos-Hernandez, R. A. Sanford, N. M. Mesbah and F. E. Loffler, Appl. Environ. Microbiol., 2006, 72, 2775–2782 CrossRef CAS PubMed.
- G. E. Fox, J. D. Wisotzkey and P. Jurtshuk, Int. J. Syst. Bacteriol., 1992, 42, 166–170 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra06211j |
| This journal is © The Royal Society of Chemistry 2015 |