Targeted killing of metastatic cells using a platelet-inspired drug delivery system
Abstract
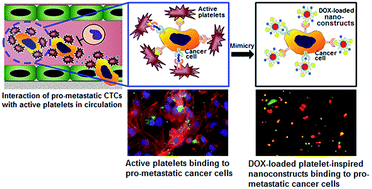
The ‘targeted nanomedicine’ approach has revolutionized cancer therapy by packaging drugs within nanovehicles that can accumulate preferentially within the tumor due to the ‘enhanced permeation and retention’ (EPR) mechanism (passive targeting) and further bind to cancer cells via specific ligand–receptor interactions (active targeting). While these approaches have shown promise in well-vascularized primary tumors, their use for metastatic sites faces challenges due to high heterogeneity in the expression levels of tumor-associated targetable receptors. Therefore, alternative strategies for metastatic cell-specific targeting are of great clinical interest. To this end, we are exploring binding interactions of metastatic cells with host cells in the vascular compartment and whether such interactions can be exploited for metastasis-targeted delivery of drug-loaded nanovehicles. For ‘proof-of-concept’ of this approach, using high-metastatic MDA-MB-231 versus low-metastatic MCF-7 human breast cancer cells, we demonstrate that high-metastatic cells have enhanced binding interactions with active platelets via several cell-surface proteins. Utilizing this information and using liposomes as a model nanovehicle, we have created delivery systems that can actively bind to the high-metastatic cells via platelet-inspired heteromultivalent interactions. Using a mono-culture of MCF-7 versus MDA-MB-231, we demonstrate that these systems enhance the delivery of Doxorubicin (DOX) to the high-metastatic cells to cause significant cell-killing. Furthermore, incubating co-cultures of MCF-7 and MDA-MB-231 cells with the DOX-loaded platelet-inspired nanovehicles, we demonstrate that the targeted binding, DOX delivery and cell-killing are quite selective towards the high-metastatic cells. Our results suggest that this platelet-inspired targeting can lead to unique strategies for metastasis-selective nanomedicine.

 Please wait while we load your content...
Please wait while we load your content...