Effect of Pb(II) on phenanthrene degradation by new isolated Bacillus sp. P1
Shao-Heng Liuab,
Guang-Ming Zeng*ab,
Qiu-Ya Niu*ab,
Ji-Lai Gongab,
Xin-Jiang Huab,
Lun-Hui Luabc,
Yao-Yu Zhouab,
Xi Huab,
Ming Chenab and
Ming Yanab
aCollege of Environmental Science and Engineering, Hunan University, Changsha 410082, PR China. E-mail: zgming@hnu.edu.cn; niuqiuya@hnu.edu.cn; Fax: +86 0731 88823701; Tel: +86 0731 88822754
bKey Laboratory of Environmental Biology and Pollution Control (Hunan University), Ministry of Education, Changsha 410082, PR China
cKey Laboratory of Reservoir Aquatic Environment, Chongqing Institute of Green and Intelligent Technology, Chinese Academy of Sciences, Chongqing 400714, China
First published on 10th June 2015
Abstract
A polycyclic aromatic hydrocarbon (PAH)-degrading microbe was isolated and the effect of Pb(II) on the degradation of phenanthrene (PHE) by this microbe was studied in this research. The changes in the metabolism, protein content, enzyme activity and PHE degradation induced by Pb(II) were investigated to elucidate the effect of Pb(II) on the degradation of PHE. The protein content, catechol 2,3-dioxygenase activity and phenanthrene-degrading ability of the enzymes were all enhanced at low concentrations of Pb(II) (below 50 mg L−1) but decreased at relatively higher concentrations (100 to 300 mg L−1) of Pb(II). These results indicate that the presence of Pb(II) is relevant to enzymatic changes in this PAH-biodegrading microbe: it helps the process of PHE biodegradation at low concentrations, but impairs PHE biodegradation at excessive concentrations.
1. Introduction
Polycyclic aromatic hydrocarbons (PAHs) can be released into the environment through nature forces or anthropogenic activities, ranging from industrial behavior to residential activities.1 They can exert a detrimental effect on both the environment and human health because of their toxicity, mutagenicity and carcinogenicity. PAHs can be removed by adsorption, volatilization, photolysis, and degradation.2,3 Amongst these methods, biodegradation is becoming a popular process due to the advantages of low cost and non-secondary pollution.4–6 Recent studies indicate that composting systems with a large biomass of microorganisms are capable of decomposing organics such as pentachlorophenol and phenolic compounds.7,8 Low molecular weight PAHs such as naphthalene (NAP), anthracene (ANT) and phenanthrene (PHE) can also be degraded by microorganisms efficiently and effectively.9,10PAHs and heavy metals prevalently occur together in contaminated environments such as manufacturing coke plants, railway yards and refinery sites, and the coexisting heavy metals may affect the PAH biodegradation.11 Niu et al.12 pointed out that Pb(II) inhibited the biodegradation of PHE in composted soil with combined pollutants PHE and Pb(II). Yang et al.13 demonstrated that Cd(II) significantly inhibited the activities of catalase, urease, phosphatase and invertase enzymes, thereby inhibiting the degradation of PAHs. Shen et al.14 found that the urease and dehydrogenase activities were far more inhibited by a synergistic effect between Zn and PHE than by individual effects. Ke et al.15 observed that metal ions posed a positive effect on the degradation of low molecular weight PAHs (fluorene (FLU) and PHE). Increasing studies have paid attention to the combined pollution of PAHs and heavy metals,16–18 although the mechanism by which heavy metals affect PAH degradation are still not deeply understood. Moreover, the enzymatic activity involved in the main process of PAH degradation may be changed by heavy metals, thus affecting the PAH degradation process. However, few studies have combined the analysis of enzymatic changes resulted from PAHs and heavy metals.
In this work, a PAH-degrading microorganism, Bacillus sp. P1, was isolated. PHE is a three-ring PAH which is often considered as a model of PAHs because it is the smallest PAH to have a “K-region” and a “bay-region”, which are the basic structures shared with other carcinogenic PAHs. Lead is widely spread in the environment and can cause severe health effects to human beings at excessive concentrations. PHE and lead were selected in this experiment as the typical and ubiquitous models of PAH and heavy metal, respectively. The objectives of this research were: (1) to investigate the effect of Pb(II) on PHE degradation by a PAH-degrading microbe; (2) to discuss the variation in protein content, activity and phenanthrene-degrading ability of enzymes extracted from the PAH-degrading microbe in the presence of Pb(II), which may impact the PHE degradation rate.
2. Materials and methods
2.1 Microorganism and medium
The bacterium was isolated with PAHs (containing fluoranthene (FLT), pyrene (PYR), PHE, and FLU) as sole carbon and energy source from the composting samples according to previous studies.12,19 A mineral medium (MnSO4 0.0447 mg L−1, ZnSO4 0.0686 mg L−1, (NH4)6MO7O24·4H2O 0.0347 mg L−1, K2HPO4 129.15 mg L−1, Na2HPO4 167 mg L−1, KH2PO4 43.5 mg L−1, NH4Cl 25 mg L−1, MgSO4 13.8 mg L−1, CaCl2 36.4 mg L−1, FeCl3 0.42 mg L−1) was used in this work.20 The molecular identification was performed by TaKaRa Biotechnology (Dalian, China). The obtained 16S rDNA sequence and similar sequences from National Center for Biotechnology Information (NCBI) were used to construct the phylogenetic tree. Branch support was assessed using 1000 bootstrap replicates.2.2 PHE biodegradation by Bacillus sp. P1
The effect of Pb(II) on the biodegradation of PHE by Bacillus sp. P1 was assessed as follows. A PHE stock solution was added to 100 mL mineral medium to obtain a final PHE concentration of 60 mg L−1. The sample group without Pb(II) was served as the control. The testing group was set with 60 mg L−1 PHE and 100 mg L−1 Pb(II).12 Both groups were sterilized before inoculation. The isolated strain with an inoculum size (OD600) of 0.8 was incubated at 30 °C and 150 rpm. Residual PHE concentration was determined daily. PHE was extracted with n-hexane before being detected with a UV-visible spectrophotometer (SHIMADZU, UV 2550) at 254 nm.21 All samples were analyzed in triplicate. The degradation rate of PHE was calculated according to the equation R = (C0 − Ct)/C0, where R is the degradation rate of PHE, C0 is the initial concentration of PHE, and Ct is the final concentration of PHE.2.3 Detection of PHE biodegradation intermediates
The effect of Pb(II) on the metabolites from PHE biodegradation was analyzed by gas chromatography-mass spectrometry (GC-MS, QP2010, Shimadzu). The isolated strain was cultured in a mineral medium system with PHE (60 mg L−1) and PHE–Pb(II) (PHE, 60 mg L−1; Pb(II), 100 mg L−1) respectively for 2 days at 30 °C and 150 rpm. The supernatant after centrifugation (15 min, 9000 rpm) was extracted in dichloromethane and then dried over anhydrous sodium sulfate before the assessment of metabolites. The injector temperature was programmed to 270 °C. Helium was used as the carrier gas at a rate of 3 mL min−1. The column temperature program was set as follows: 120 °C for 1 min, and then 3 °C min−1 up to 280 °C.222.4 Extraction of extracellular enzymes and intracellular enzymes
The centrifuged supernatants (9000 rpm, 4 °C) after culturing for 4 days in mineral medium were extracellular proteins, which were approximately considered as crude extracellular enzymes (EE). The residues were washed with phosphate buffer (0.05 mM, pH 6.8) three times and then sonicated (300 W, 3 s/8 s) for 10 min at 4 °C, followed by subsequent centrifugation (9000 rpm, 4 °C). The supernatants were intracellular proteins, which can similarly be seen as crude intracellular enzymes (IE).232.5 Analysis of extracellular and intracellular protein content
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) fingerprint analysis was carried out to investigate the effects of Pb(II) on the protein composition extracted from the PAH-degrading microbe. SDS-PAGE was performed according to the protocol of Laemmli24 with 12% polyacrylamide gel, using a mini-protein vertical electrophoresis system (Bio-Rad, USA). The samples were secretase-extracted from the bacteria cultivated in PHE (60 mg L−1) and PHE–Pb(II) (PHE, 60 mg L−1; Pb(II), 50, 100, 150, 200, 300 mg L−1) media, respectively. They were salted and dialyzed before loading into the gel. SDS-PAGE gel was stained with Coomassie brilliant blue R-250 and the molecular mass of the samples was determined by the relative mobility of the stained SDS-PAGE marker running alongside.2.6 Determination of catechol 2,3-dioxygenase activity
Catechol 2,3-dioxygenase is the key enzyme involved in the degradation of aromatic hydrocarbons.25,26 Crude extracellular and intracellular enzymes were extracted from the PAH-biodegrading microbe cultivated in PHE (60 mg L−1) and PHE–Pb(II) (PHE, 60 mg L−1; Pb(II), 50, 100, 150, 200, and 300 mg L−1) media for 2 days at 30 °C and 150 rpm, respectively, for the detection of catechol 2,3-dioxygenase. The mixtures containing 2.4 mL phosphate buffer (0.05 mM), 0.4 mL catechol (20 μM) and 0.2 mL EE (or IE) were analyzed for the activity of catechol 2,3-dioxygenase at 375 nm by UV-visible spectrophotometry. One unit of activity was defined as 1 μmol of substrate converted per minute. The soluble protein content was measured following the procedure described by Bradford.272.7 Assessment of Pb(II) on enzymes extracted from the PAH-biodegrading microbe
Conditions of pH and temperature were varied for a thorough understanding of the effect of Pb(II) on PHE degradation. Experiments were conducted in five Erlenmeyer flasks each containing 20 mL ultrapure water and 25 mg L−1 of PHE. The pH value was adjusted to 5.0, 6.0, 7.0, 8.0, and 9.0 with HCl or NaOH, respectively. Enzymes (2 mL) were added to each flask and then shaken for 30 min at 30 °C and 150 rpm. Moderate temperatures were set as 25, 30 and 37 °C, respectively. A group with enzymes which had been inactivated at high temperature was used as the control for enzymatic biodegradation of PHE.To investigate the degradation of PHE by enzymes, 2 mL enzyme solution was added into 50 mL ultrapure water containing 5 mg L−1 PHE, followed by the addition of various concentrations of Pb(II) (0, 50, 100, 150, 200, and 300 mg L−1). Flasks were incubated at 30 °C and agitated at 150 rpm. PHE was extracted and determined after 30 min as per the method described in Section 2.2.
2.8 Statistical analysis
Statistically significant differences between the results were evaluated on the basis of standard deviation determinations and one-way analysis of variance (ANOVA, 95% confidence intervals test) using the SPSS 19.0 software.3. Results and discussion
3.1 Effect of Pb(II) on the PHE biodegradation by PAH-biodegrading microbe
The bacteria isolated from the compost belongs to the genus Bacillus and the highest homology obtained by this strain was 100% of similarity to Bacillus malacitensis. The 16S rDNA sequence was submitted to the GenBank and given the accession number KP888558. The phylogenetic tree based on a multiple sequence alignment of the 16S rDNA sequence is presented in Fig. 1. This strain was named Bacillus sp. P1.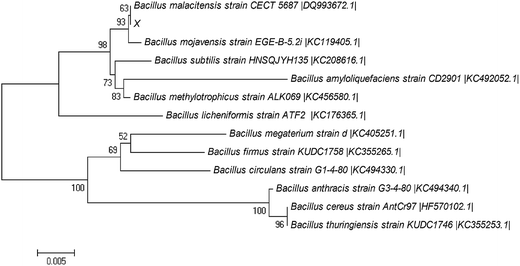 | ||
| Fig. 1 Phylogenetic tree established by the neighbor-joining method based on the 16S rDNA sequence of Bacillus sp. P1 and the similar sequences obtained from NCBI. | ||
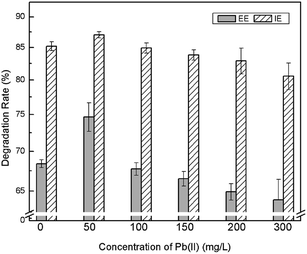
Bacillus sp. P1 presents satisfactory potential in degrading PHE according to Fig. 2. PHE was degraded significantly (p < 0.05) in both media (with and without Pb(II)) in the first two days. In this period, the degradation rate in PHE (60 mg L−1) and PHE–Pb(II) (PHE, 60 mg L−1; Pb(II), 100 mg L−1) media rapidly increased to 91.9% and 85.7%, respectively. The degradation rate of PHE in the medium with Pb(II) was higher than that in the medium without Pb(II) on the first day, mainly due to the increased adsorption of PHE by Bacillus sp. P1, since Pb(II) might enhance the formation of Bacillus sp. P1–Pb(II)–PHE ternary surface complexes.28,29 Bacillus sp. P1 began to grow logarithmically on day 2 and promoted a higher rate of PHE degradation. During the 6 day period incubation, PHE degradation reached a plateau in the two groups at a degradation rate of 95.1%. The degradation rate of PHE in the medium with Pb(II) was lower compared to that in the medium without Pb(II) from day 2 to 6, because Pb(II) hindered the decay of PHE, which might be attributed to the inhibiting effect of Pb(II) over Bacillus sp. P1 resulting in lower degradation rates of PHE. Bacillus sp. P1 can be affected by Pb(II) for the following three reasons: (1) the motor function of the cytoskeletons may be hindered by Pb(II), which may consequently retard the cytokinesis in Bacillus sp. P1;30 (2) Pb(II) may enhance the oxidation process produced by reactive oxygen species (ROS), thereby resulting in further damage to the cell organelles;15,31 (3) Pb(II) may penetrate through the cell wall of the bacteria oxidizing the surface proteins on the plasma membrane directly and consequently disturbing the cell homeostasis.32,33
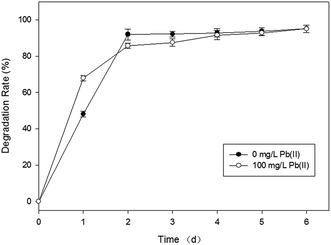 | ||
| Fig. 2 Effect of Pb(II) on PHE biodegradation by Bacillus sp. P1 with respect to the incubation time at 30 °C. The error bars represent ±1 standard deviation. | ||
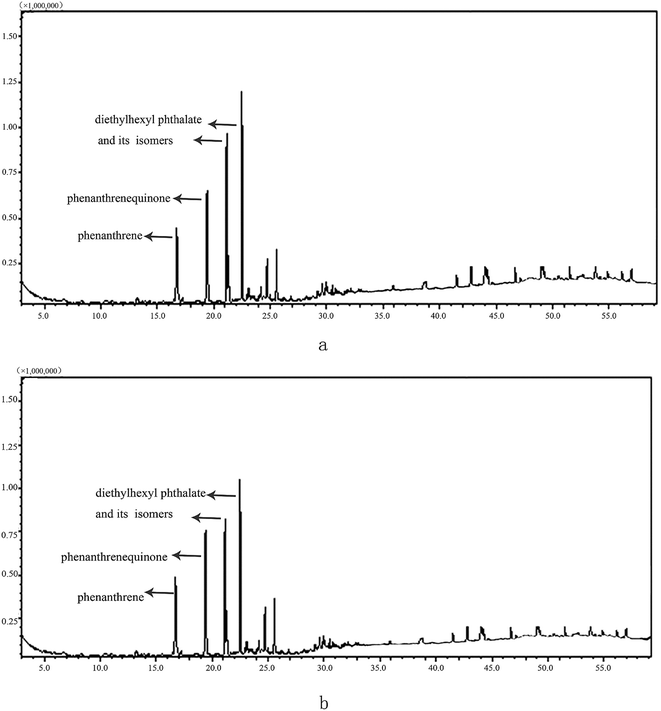
The intermediates of PHE were investigated in this study according to Fig. 3. Qualitative analysis was based on retention indexes, and mass spectrum comparison with data in the NIST08 library. The major metabolites detected in both PHE and PHE–Pb(II) systems were diethylhexyl phthalate (m/z 93, 104, 122, 149, 167, 205, 223) and phenanthrenequinone (m/z 76, 152, 180, 208).34 The results suggest that Bacillus sp. P1 degrades PHE via a protocatechuate pathway.
3.2 Effect of Pb(II) on protein content of Bacillus sp. P1
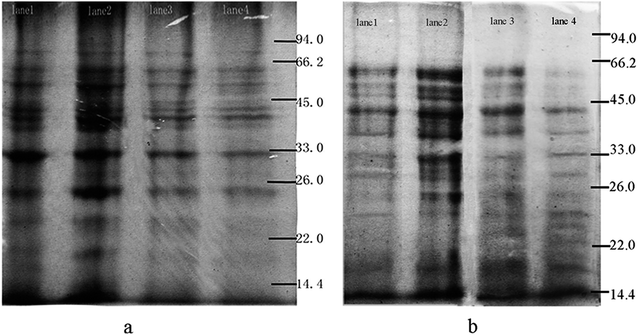
The alteration of the extracellular protein (EP) and intracellular protein (IP) concentrations were analyzed through the SDS-PAGE fingerprint with the Gelpro software (Fig. 4). With the increase of Pb(II) concentration from 0 to 50 mg L−1, the total content (absolute integrated optical density) of EP and IP increased from 5.8 × 105 to 6.4 × 105, and from 7.2 × 105 to 7.5 × 105, respectively, and then declined from 4.2 × 105 to 4.1 × 105, and from 7.1 × 105 to 5.3 × 105 respectively with the increase of Pb(II) concentration from 100 to 150 mg L−1. The change in the expression of total protein might be attributed to the gene clusters involved in the degradation of PHE being induced by Pb(II). Besides, hormesis induced by Pb(II) can cause an increase in protein amount, which protects the cells from the toxic effect of Pb(II) by exporting the cation. The proteins finally could be reduced due to the toxic effect of Pb(II) on Bacillus sp. P1.35 The protein bands with molecular mass of 33–45 kDa of EP (IP) increased from 8.6 × 104 to 1.3 × 105 (from 1.7 × 105 to 2.5 × 105), then decreased from 8.3 × 104 to 7.2 × 105 (from 1.4 × 105 to 1.2 × 105). It is probable that a low concentration of Pb(II) stimulates the release of the key ring-opening enzymes, thus promoting the degradation rate of PHE.3.3 Effect of Pb(II) on catechol 2,3-dioxygenase activity
Table 1 lists the catechol 2,3-dioxygenase activity of EE and IE in the presence of different concentrations of Pb(II) during the process of PHE degradation. The results show a remarkable variation (p < 0.05) in the catechol 2,3-dioxygenase activity of EE and IE in the presence of Pb(II) during PHE degradation. The catechol 2,3-dioxygenase activity of EE showed a circuitous profile in the presence of Pb(II). The activity increased in the presence of a low concentration of Pb(II) (below 50 mg L−1), but conversely dropped down in the presence of a high concentration of Pb(II) (above 100 mg L−1). For IE, the catechol 2,3-dioxygenase activity increased with the increasing Pb(II) concentration and reached a maximum in the system at 50 mg L−1 Pb(II), then declined as the Pb(II) concentration increased. These results are consistent with the results shown in Fig. 4, indicating that catechol 2,3-dioxygenase is involved in the degradation of PHE due to the positive correlation between PHE utilization and the activity of catechol 2,3-dioxygenase in the presence of low concentrations of Pb(II). It might be possible that enzyme activities are inhibited by Pb(II) by denaturing the protein structure or masking catalytically active groups of enzymes.36 The latter reduction of catechol 2,3-dioxygenase activity may be ascribed to the direct suppression of microbial growth by Pb(II).| Concentration of Pb(II) (mg L−1) | Content of protein (μg) | Specific activity of catechol 2,3-dioxygenase (U mg−1) | ||
|---|---|---|---|---|
| Extracellular enzymes | Intracellular enzymes | Extracellular enzymes | Intracellular enzymes | |
| 0 | 132.5 | 117.5 | 274.6 | 890.1 |
| 50 | 186.7 | 133.3 | 296.1 | 1003.3 |
| 100 | 131.7 | 112.7 | 244.0 | 784.9 |
| 150 | 129.2 | 80.5 | 203.8 | 760.2 |
| 200 | 102.6 | 73.9 | 133.4 | 413.1 |
| 300 | 95.3 | 70.6 | 99.3 | 304.5 |
3.4 Effect of Pb(II) on PHE degradation by enzymes
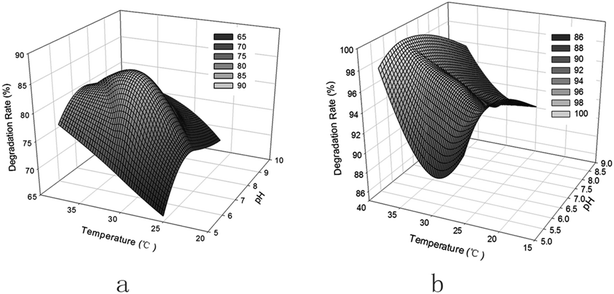 | ||
| Fig. 5 Effect of pH and temperature on PHE degradation by extracellular enzymes (a) and intracellular enzymes (b). | ||
PHE degradation by EE and IE increases with the increase in temperature from 25 to 37 °C, the degradation rates by EE and IE being 71.8% and 99.8% at 37 °C, respectively. A higher temperature promotes the enzymatic activity. Besides, the solubility of PHE would increase with the increase of temperature, therefore leading to a higher bioavailability governed by solubility.38 IE displayed a higher degradation ability than EE, which could be attributed to the fact that more dioxygenase enzymes involved in the steps of PHE catabolism, such as naphthalene 1,2-dioxygenase, are present in IE.39
4. Conclusions
Low concentrations of Pb(II) have a positive effect on PHE degradation, while high concentrations of Pb(II) have the opposite effect on the degradation of PHE. Protein content, the activity of catechol 2,3-dioxygenase and PHE degradation by enzymes were all gently enhanced under exposure to relatively low concentrations of Pb(II) due to the formation of enzyme–metal–substrate complexes, and all three indexes were inhibited by high concentrations of Pb(II) because of the metal’s influence on the enzymes of the PAH-degrading microbe. The aforementioned results suggest that Pb(II) could impact PHE biodegradation by changing the protein content, enzyme activity and phenanthrene-degrading ability of enzymes secreted by Bacillus sp. P1, which contribute to the understanding of the pollution mechanism by combined heavy metals and PAHs.Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (51108166, 51378190) and the Program for Changjiang Scholars and Innovative Research Team in University (IRT-13R17).References
- P. M. Mwanamoki, N. Devarajan, F. Thevenon, N. Birane, L. F. de Alencastro, D. Grandjean, P. T. Mpiana, K. Prabakar, J. I. Mubedi, C. G. Kabele, W. Wildi and J. Poté, Chemosphere, 2014, 111, 485–492 CrossRef CAS PubMed.
- J. M. DeBruyn, T. J. Mead and G. S. Sayler, Environ. Sci. Technol., 2012, 46, 99–106 CrossRef CAS PubMed.
- H. Lee, Y. Jang, Y. S. Choi, M. J. Kim, J. Lee, H. Lee, J. H. Hong, Y. M. Lee, G. H. Kim and J. J. Kim, J. Microbiol. Methods, 2014, 97, 56–62 CrossRef CAS PubMed.
- I. Vitte, R. Duran, R. Jézéquel, P. Caumette and C. Cravo-Laureau, Environ. Sci. Pollut. Res., 2011, 18, 1022–1032 CrossRef CAS PubMed.
- G. Zeng, M. Chen and Z. Zeng, Science, 2013, 340, 1403 CrossRef CAS PubMed.
- Y. Liu and X. Hua, RSC Adv., 2014, 4, 31120–31122 RSC.
- G. Zeng, Z. Yu, Y. Chen, J. Zhang, H. Li, M. Yu and M. Zhao, Bioresour. Technol., 2011, 102, 5905–5911 CrossRef CAS PubMed.
- Y. Liu, Z. Zeng, G. Zeng, L. Tang, Y. Pang, Z. Li, C. Liu, X. Lei, M. Wu, P. Ren, Z. Liu, M. Chen and G. Xie, Bioresour. Technol., 2012, 115, 21–26 CrossRef CAS PubMed.
- Z. Khan, D. Roman, T. Kintz, M. delas Alas, R. Yap and S. Doty, Environ. Sci. Technol., 2014, 48, 12221–12228 CrossRef CAS PubMed.
- R. Xiao, J. Bai, J. Wang, Q. Lu, Q. Zhao, B. Cui and X. Liu, Chemosphere, 2014, 110, 8–16 CrossRef CAS PubMed.
- S. Murthy, G. Bali and S. K. Sarangi, J. Environ. Biol., 2013, 35, 407–411 Search PubMed.
- Q. Niu, G. Zeng, Y. Niu and F. Zeng, ICBBE 2009. 3rd International Conference on Bioinformatics and Biomedical Engineering, IEEE, Beijing, June, 11–13, 2009 Search PubMed.
- Z. X. Yang, S. Q. Liu, D. W. Zheng and S. D. Feng, J. Environ. Sci., 2006, 18, 1135–1141 CrossRef CAS.
- G. Shen, Y. Lu, Q. Zhou and J. Hong, Chemosphere, 2005, 61, 1175–1182 CrossRef CAS PubMed.
- L. Ke, L. Luo, P. Wang, T. Luan and N. F. Y. Tam, Bioresour. Technol., 2010, 101, 6950–6961 CrossRef CAS PubMed.
- B. A. M. Bandowe, M. Bigalke, L. Boamah, E. Nyarko, F. K. Saalia and W. Wilcke, Environ. Int., 2014, 65, 135–146 CrossRef CAS PubMed.
- R. Kamalakkannan, V. Zettel, A. Goubatchev, K. Stead-Dexter and N. I. Ward, J. Environ. Monit., 2004, 6, 175–181 RSC.
- L. Sun, X. Liao, X. Yan, G. Zhu and D. Ma, Environ. Sci. Pollut. Res., 2014, 21, 12494–12504 CrossRef CAS PubMed.
- I. Ghosh, J. Jasmine and S. Mukherji, Bioresour. Technol., 2014, 166, 548–558 CrossRef CAS PubMed.
- F. Moscoso, I. Teijiz, F. J. Deive and M. A. Sanromán, Bioresour. Technol., 2012, 119, 270–276 CrossRef CAS PubMed.
- N. D. Marsh, C. J. Mikolajczak and M. J. Wornat, Spectrochim. Acta, Part A, 2000, 56, 1499–1511 CrossRef.
- A. Janbandhu and M. H. Fulekar, J. Hazard. Mater., 2011, 187, 333–340 CrossRef CAS PubMed.
- T. G. Luan, K. S. H. Yu, Y. Zhong, H. W. Zhou, C. Y. Lan and N. F. Y. Tam, Chemosphere, 2006, 65, 2289–2296 CrossRef CAS PubMed.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
- D. Wojcieszyńska, K. Hupert Kocurek, I. Greń and U. Guzik, Int. Biodeterior. Biodegrad., 2011, 65, 853–858 CrossRef PubMed.
- S. N. Singh, B. Kumari, S. K. Upadhyay, S. Mishra and D. Kumar, Bioresour. Technol., 2013, 133, 293–300 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- C. Anyika, Z. Abdul Majid, Z. Ibrahim, M. Zakaria and A. Yahya, Environ. Sci. Pollut. Res., 2015, 22, 3314–3328 CrossRef CAS PubMed.
- J. Zhao and M. C. He, Appl. Surf. Sci., 2014, 317, 718–723 CrossRef CAS PubMed.
- H. L. Su, C. C. Chou, D. J. Hung, S. H. Lin, I. C. Pao, J. H. Lin, F. L. Huang, R. X. Dong and J. J. Lin, Biomaterials, 2009, 30, 5979–5987 CrossRef CAS PubMed.
- L. Lei, A. P. Khodadoust, M. T. Suidan and H. H. Tabak, Water Res., 2005, 39, 349–361 CrossRef CAS PubMed.
- J. Wang and C. Chen, Biotechnol. Adv., 2009, 27, 195–226 CrossRef CAS PubMed.
- W. W. Tang, G. M. Zeng, J. L. Gong, J. Liang, P. Xu, C. Zhang and B. B. Huang, Sci. Total Environ., 2014, 468–469, 1014–1027 CrossRef CAS PubMed.
- K. Masakorala, J. Yao, M. Cai, R. Chandankere, H. Yuan and H. Chen, J. Hazard. Mater., 2013, 263(2), 493–500 CrossRef CAS PubMed.
- H. Guo, S. Luo, L. Chen, X. Xiao, Q. Xi, W. Wei, G. Zeng, C. Liu, Y. Wan, J. Chen and Y. He, Bioresour. Technol., 2010, 101, 8599–8605 CrossRef CAS PubMed.
- L. S. Zeng, M. Liao, C. L. Chen and C. Y. Huang, Ecotoxicol. Environ. Saf., 2007, 67, 67–74 CrossRef CAS PubMed.
- M. Dussault, V. Bécaert, M. François, S. Sauvé and L. Deschênes, Chemosphere, 2008, 72, 755–762 CrossRef CAS PubMed.
- S. M. Bamforth and I. Singleton, J. Chem. Technol. Biotechnol., 2005, 80, 723–736 CrossRef CAS PubMed.
- S. C. Corgie, F. Fons, T. Beguiristain and C. Leyval, Mycorrhiza, 2006, 16, 207–212 CrossRef CAS PubMed.
- J. V. Joseph and A. S. Mildvan, J. Biol. Chem., 1971, 247, 3454–3563 Search PubMed.
- A. Karaca, S. C. Cetin, O. C. Turgay and R. Kizilkaya, Soil heavy metals, Springer, 2010, vol. 19, ch. 4, pp. 237–262 Search PubMed.
| This journal is © The Royal Society of Chemistry 2015 |