Amylosucrase-mediated synthesis and self-assembly of amylose magnetic microparticles†
Min-Cheol Lim,
Gwan-Hyung Lee,
Duyen Thi Ngoc Huynh,
Carlos Andres Morales Letona,
Dong-Ho Seo,
Cheon-Seok Park and
Young-Rok Kim*
Graduate School of Biotechnology & Department of Food Science and Biotechnology, Kyung Hee University, Yongin 446-701, Korea. E-mail: youngkim@khu.ac.kr; Fax: +82 31 204 8116; Tel: +82 31 201 3830
First published on 15th April 2015
Abstract
This paper reports a one-step bottom-up approach for the preparation of amylose magnetic beads (AMBs) via enzymatic synthesis and a self-assembly process of amylose and iron oxide nanoparticles. The resulting AMBs were highly effective in the column free purification of target protein with high binding capacity and recyclability.
Surface functionalized magnetic beads are the key materials for simple and small volume protein purification because they eliminate the need for expensive liquid chromatography systems, centrifuges, filters or other equipment.1 Selective separation and purification of the target proteins from complex matrices is essential for characterizing the function, structure and interaction of proteins. A number of ligands capable of binding to proteins with very high affinity and selectivity have attracted considerable attention over the past few decades for applications in the separation and purification of proteins.2 The main challenges in this field are achieving a high grafting density of ligands on the surface of the resin and excellent magnetic characteristics to increase the binding capacity and purification efficiency. Several studies have focused on the development of a range of magnetic particles with enhanced dispersibility, biocompatibility and surface functionality suitable for a range of downstream applications. On the other hand, few have reported the preparation of magnetic particles for the separation and purification of maltose binding protein (MBP)-tagged proteins.3 MBP has a specific affinity to amylose, which is a repeating maltose polymer with flexible polysaccharide chains joined by α-(1 → 4) links. MBP is encoded by the malE gene of Escherichia coli K12 and has been used to simplify the purification of the target protein by its affinity to cross-linked amylose. MBP-tagging is particularly useful in protein purification because it increases the solubility and resistance to intracellular proteolysis of the target proteins.4 For the preparation of magnetic beads coated with affinity ligands, such as amylose or maltodextrin, functionalization of the surface often requires multistep procedures, pretreatment, cleaning, and crosslinking chemistry.5 Moreover, the purification capacity of the magnetic beads is influenced predominantly by the grafting density of the specific ligand functionalized on the surface of the platform materials.6
This paper reports a facile and efficient approach to the synthesis of amylose magnetic microbeads (AMB) consisting of pure amylose molecules and iron oxide nanoparticles through the unique catalytic characteristics of amylosucrase (ASase) in the presence of sucrose and iron oxide magnetic nanoparticles. ASase from Deinococcus geothermalis (DGAS) was reported to catalyze the synthesis of the amylose chain from sucrose, and the resulting amylose chain self assembles into a spherical microstructure in the same reaction.7
In this study, it was demonstrated that iron oxide nanoparticles are efficiently incorporated into the amylose microbeads during the self-assembly process, and the resulting AMBs were found to be excellent in magnetic separation and purification of the MBP-tagged proteins. The efficiency of AMBs as affinity templates for the magnetic separation and purification of proteins has been demonstrated using the MBP-tagged green fluorescent protein (GFP) as a model protein. The binding capacity, selectivity and recyclability of the AMBs were examined to evaluate their potential applications in the separation of biomolecules.
Fig. 1a presents the overall scheme for the synthesis of AMBs via a one-step enzymatic reaction using ASase. The polymerization of amylose molecules was initiated by the hydrolysis of sucrose to fructose and glucose by ASase. The free glucose molecules were polymerized to malto-oligosaccharide and a longer amylose chain by the same enzyme, which were then self-associated to form a spherical bead structure via a hydrophobic interaction and hydrogen bonding.8 The hydrophobic guest agent, an iron oxide nanoparticle, was incorporated efficiently into the amylose structure during the enzymatic synthesis and self-assembly process of the amylose molecules. The enzymatic synthesis was carried out in a 1 ml reaction volume containing 500 mM sucrose, 300 U DGAS and 200 μl of iron oxide nanoparticle solution (0.8–1.4% (w/v) in heptane) at 30 °C for 24 h. The reaction mixture was bath sonicated prior to the addition of the enzyme to disperse the organic solvent in the aqueous reaction solution and placed on angle adjustable rotator during the entire polymerization reaction. The resulting AMBs were shown to have a well-defined spherical shape (Fig. 1b), suggesting that the enzymatically synthesized amylose molecules were self-assembled spontaneously into spherical micro-sized beads with iron oxide nanoparticles embedded within the amylose microbeads. The mean size of the AMBs was determined to be 2.09 ± 0.42 μm (Fig. S2†). As shown in Fig. 1b, the synthesized AMB was brown due to the embedded iron oxide nanoparticles, whereas the control reaction without iron oxide nanoparticles resulted in a white product (Fig. S1a†).
Fig. 2a–d shows a TEM image and electron maps of carbon, oxygen and Fe in the AMBs. The TEM image and elemental mapping data clearly show iron oxide nanoparticles distributed evenly within the spherical amylose bead. The carbon and oxygen in AMB are derived from the amylose molecules. As shown in XRD pattern (Fig. 2e), the amylose beads (iii) showed diffraction peaks at 15, 17, 22, and 24° 2θ, which are characteristic of the pure B-type crystal structure of amylose.9 The iron oxide nanoparticles (i) have a typical XRD pattern of Fe3O4 with peaks at 30, 35, 43, 57, and 62° 2θ.10 The AMBs (ii) showed the all representative peaks for amylose and iron oxide nanoparticles. The amplitude of the peaks for AMB were lower than those of the pure amylose beads suggesting that the crystallinity of amylose in AMB was disrupted slightly by the embedded iron oxide nanoparticles.
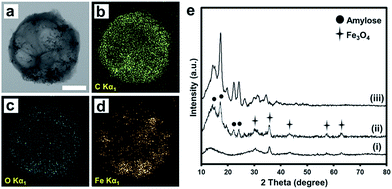 | ||
| Fig. 2 (a) TEM image and elemental mapping of carbon, oxygen and Fe (b–d) in AMB, scale bar is 500 nm. (e) XRD patterns of (i) iron oxide nanoparticle, (ii) AMBs, and (iii) amylose beads. | ||
The magnetic properties of AMBs and pure amylose beads were examined using a vibrating sample magnetization (VSM) magnetometer. The hysteresis loop data exhibited strong superparamagnetic behavior of AMB with a saturated magnetization value of 4.5 emu g−1, whereas the pure amylose beads showed virtually no magnetic property (Fig. 3a). The AMBs could be dispersed in water and were readily separated from their homogeneous suspension by the external magnetic field. In addition, the aggregated amylose magnetic beads were resuspended easily by simple shaking after removing the magnetic field (Fig. 3b). These results confirm that the prepared AMBs have excellent magnetic responsibility and redispersibility for actual applications in magnetic separation.
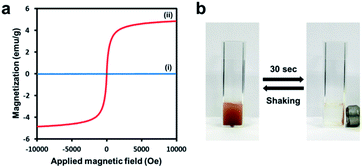 | ||
| Fig. 3 (a) Magnetic hysteresis loops of (i) pure amylose beads and (ii) AMBs. (b) The separation of AMBs dispersed in water by magnet in 30 s. | ||
To verify the potential applications in magnetic separation and protein purification, the AMBs were tested to purify MBP-tagged GFP from the cell lysate of E. coli overexpressing the target protein. The expression vector was constructed by ligating the gfp gene into the pMal-c2X vector, and transferring it to E. coli DH5α. The expression of the target protein from E. coli DH5α harboring the vector was induced with 0.1 mM IPTG, followed by growing the host strain overnight at 18 °C. The cells were harvested by centrifugation and lysed by ultrasonication in a lysis buffer (ESI†). The resulting 1 ml lysate was incubated with 5 mg of AMB at 4 °C for 30 min and the MBP-tagged GFP captured on the AMB was then separated by a magnet. Fig. 4 shows the magnetic separation of the MBP-tagged GFP using AMB. The MBP-tagged GFPs were effectively separated together with AMB but the GFPs without the MBP-tag remained in solution. The collected AMBs were washed three times with water and observed by fluorescence microscopy (Fig. 4c–d). The surface of the AMBs was observed as green due to the bound MBP-tagged GFP. On the other hand, no green fluorescence was observed in the AMBs that had reacted with GFP without a MBP-tag, which suggests that binding is through the specific interaction between amylose and the MBP-tag.
The captured MBP-tagged GFPs can be released from the surface of AMBs using the elution buffer containing 10 mM maltose. The free maltose molecule competes with the AMBs for the binding site of MBP. Therefore, excessive maltose is effective in releasing all the MBP-tagged protein from the amylose beads. All the fractions during protein purification were analyzed by SDS-PAGE (Fig. 5a). The results show that AMB is an effective material for purifying the MBP-tagged protein. The strong and selective affinity of the AMB to MBP-tag enabled purification of the target protein (MBP-tagged GFP, 72 kDa) with a high yield and purity. The purification capacity of AMB for MBP-tagged GFP was determined to be 72.31 μg per mg of AMBs, which is more than three-fold higher than that of commercial amylose magnetic beads. The enhanced purification capacity of the enzymatically-synthesized AMB developed in this study might be result from the high surface coverage of the specific ligand. Conventional amylose magnetic beads are normally produced by the covalent attachment of specific ligands, such as maltodextrin and maltose, onto the surface of the paramagnetic beads.3 The AMBs developed in this study, however, consisted of enzymatically synthesized pure amylose molecules as the matrix materials and embedded iron oxide nanoparticles. This approach can eliminate the complicated cross-linking process needed to conjugate a specific ligand to the surface of a magnetic bead, thereby enhance the purification capacity of the MBP-tagged proteins.
To test the recyclability of AMBs, the magnetic beads were rinsed with 300 μl of water, 300 μl of 0.1% SDS, 100 μl of water, and 500 μl of column buffer to wash out the excess maltose and clean the surface of the beads prior to reuse. From the Bradford assay, the AMB maintained a purification capacity of 88%, even at the third recycle (Fig. 5b). A slight decrease in purification capacity in the recycled AMB could be attributed to the loss of AMB during the washing step between each experiment. These results highlight the specific affinity and high purification capacity of AMB. The robust nature of AMB also enabled its use multiple times without significant loss of its purification ability.
In conclusion, this paper reported a novel method for the one-step preparation of AMBs by enzymatic synthesis and the self-assembly of amylose and iron oxide nanoparticle. The AMBs had a well-defined spherical shape with a diameter of 2.09 ± 0.42 μm, which exhibit excellent magnetic responses and dispersibility in aqueous solutions. The AMB possesses a high specificity and purification capacity (72 μg mg−1) for the MBP-tagged protein as well as stability for multiple usage by simple washing. The unique catalytic characteristics of biological systems were used to synthesize amylose-based magnetic microstructure in a very simple and cost-effective manner through an enzymatic polymerization and self-assembly process. In addition to the protein purification presented in this study, this biological approach to synthesize an amylose based composite material is expected have a number of potential applications in the field of chromatography, biosensors and smart delivery of active compounds in the near future.
Acknowledgements
This study was supported by the Korea Research Foundation Grant (NRF-2013R1A1A2061841), the Pioneer Research Center Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (2012-0009575) and Korea Small and Medium Business Administration (S2176081). Dr SangYoon Park (Advanced Institutes of Convergence Technology, Seoul National University) is greatly appreciated by the authors for VSM analysis and meaningful discussions.Notes and references
- I. Safarik and M. Safarikova, Biomagn Res Technol., 2004, 2, 7 CrossRef PubMed.
- C. L. Young, Z. T. Britton and A. S. Robinson, Biotechnol. J., 2012, 7, 620–634 CrossRef CAS PubMed.
- L. Zhou, J. Wu, H. Zhang, Y. Kang, J. Guo, C. Zhang, J. Yuan and X. Xing, J. Mater. Chem., 2012, 22, 6813–6818 RSC; J. Zheng, C. Ma, Y. Sun, M. Pan, L. Li, X. Hu and W. Yang, ACS Appl. Mater. Interfaces, 2014, 6, 3568–3574 Search PubMed.
- P. Duplay and M. Hofnung, J. Bacteriol., 1988, 170, 4445–4450 CAS.
- E. Jo, M.-C. Lim, H.-N. Kim, H.-J. Paik, Y.-R. Kim and U. Jeong, J. Polym. Sci., Part B: Polym. Phys., 2011, 49, 89–95 CrossRef CAS PubMed.
- B. V. Bhut, K. A. Christensen and S. M. Husson, J. Chromatogr. A, 2010, 1217, 4946–4957 CrossRef CAS PubMed.
- M.-C. Lim, D.-H. Seo, J.-H. Jung, C.-S. Park and Y.-R. Kim, RSC Adv., 2014, 4, 26421–26424 RSC.
- A. Buleon, G. Veronese and J.-L. Putaux, Aust. J. Chem., 2007, 60, 706–718 CrossRef CAS.
- G. Potocki-Veronese, J.-L. Putaux, D. Dupeyre, C. Albenne, M. Remaud-Siméon, P. Monsan and A. Buleon, Biomacromolecules, 2005, 6, 1000–1011 CrossRef CAS PubMed.
- X. Li, H. Li, G. Liu, Z. Deng, S. Wu, P. Li, Z. Xu, H. Xu and P. K. Chu, Biomaterials, 2012, 33, 3013–3024 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures including enzymatic synthesis of amylose magnetic beads, characterization of amylose magnetic beads, purification of MBP-tagged proteins, and cloning of MBP-His-tagged GFPs. See DOI: 10.1039/c5ra02284c |
| This journal is © The Royal Society of Chemistry 2015 |



