Semi-synthesis and anti-lung cancer activity evaluation of aryl dihydrothiazol acyl podophyllotoxin ester derivatives†
Hong-Yan Linab,
Li-Fei Baic,
Fang Wangab,
Xun Wuab,
Lu-Jing Hanab,
Shahla Karim Balochab,
Yong-Hua Yang*ab and
Xiao-Ming Wang*ab
aState Key Laboratory of Pharmaceutical Biotechnology, NJU-NJFU Joint Institute of Plant Molecular Biology, Nanjing University, Nanjing, 210023, China. E-mail: Yangyh@nju.edu.cn; Wangxm07@nju.edu.cn; Fax: +86-25-89681381; Tel: +86-25-89681381
bCo-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, 210037, China
cSchool of Life Sciences and Chemistry, Jiangsu Second Normal University, Nanjing 210013, China
First published on 12th March 2015
Abstract
Lung cancer is the leading cause of cancer death worldwide, making it one of the biggest concerns for chemoprevention. In this study, we obtained seventeen potent anticancer agents through semi-synthesis based on a natural product, podophyllotoxin. Despite prior studies of podophyllotoxin derivatives being focussed on DNA-topoisomerase II, we now turn our attention to their effect on tubulin. The MTT assay screened out the most potent anticancer agent, S12 (IC50 = 0.18 μM against A549 cell line), and it showed lower cytotoxicity against normal cells. Next, the flow cytometry analysis result demonstrated that it can cause a remarkable cell cycle arrest at G2/M phase but the effect on apoptosis is not very significant. In addition, docking simulation results showed that S12 can nicely bound to the colchicine binding site of tubulin. Furthermore, we confirmed that S12 can really inhibit tubulin polymerization through confocal microscopy and protein expression determination assay.
Introduction
Lung cancer is one of the malignant tumors which have the fastest growing morbidity and mortality and pose the biggest threat to people’s health and life.1,2 Histopathologically, it is classified as a small-cell carcinoma and the non-small cell carcinoma types of adenocarcinoma, squamous-cell carcinoma, and large-cell undifferentiated carcinoma.3 Lung cancer is currently the leading cause of cancer deaths in both men and women worldwide, with over a million deaths annually and a dismal 16% five-year survival rate.4,5Tobacco smoking is the most predominant risk factor for the disease, and accounts for about 87% of lung cancer cases.6 Carcinogens present in tobacco smoke might bond covalently to DNA at certain specific sites, forming bulky adducts, creating a microenvironment that facilitates oncogenic mutations and lung cancer initiation and promotion.7 Additionally, a fraction of lung cancers arise in patients who have never smoked. Apart from a hereditary factor, the main reason is long-term exposure to a polluted environment, including second-hand smoke and increasing serious air pollution.8 In consideration of the seriousness of lung cancer, we should not only protect the environment but endeavour to prepare a new drug that is more effective and less toxic for lung cancer patients.
Natural products have long been an important source of treatments for cancer.9 At present, there are hundreds of plants that have been found to possess significant antitumor properties.10 Podophyllotoxin (1) (Fig. 1), a naturally occurring cyclolignan which is the main component of podophyllum resin, shows strong cytotoxic activity against various cancer cell lines but failed to be used in human clinical trials because of unacceptable toxic side-effects.11–14 In order to overcome the limitations, numerous researchers focus on structural modification at the 4 position of cycloparaffin (C ring) to generate derivatives with superior pharmacological profiles.15–17 Etoposide (2) and teniposide (3), two semi-synthetic epipodophyllotoxin derivatives, are successful in clinical use as antitumor agents by inhibiting the enzyme DNA-topoisomerase II.18,19 However, reports on podophyllotoxin derivatives, which exhibit antitumor activities by inhibiting cancer cellular microtubule assembly, are rare. In this study, we aim to discover some more potent and selective drugs targeting tubulin based on the podophyllotoxin scaffold.
In order to reduce the toxic side effect of podophyllotoxin itself and improve drug targeting, we plan to introduce an aryl dihydrothiazol moiety, which can improve the water solubility and pharmacokinetic parameters of the drug into the podophyllotoxin skeleton.20 Some researchers have indicated that aryl dihydrothiazole compounds can also disrupt tubulin polymerization, therefore inhibiting the production of functional microtubules and cell mitosis.21 For this purpose, we synthesized a series of novel aryl dihydrothiazol acyl podophyllotoxin ester derivatives and evaluated their anticancer activities. We hope our study will pave the way for exploring new anticancer mechanisms of podophyllotoxin derivatives.
Results and discussion
The synthetic routes for the novel aryl dihydrothiazol acyl podophyllotoxin ester derivatives S1–S17 are outlined in Scheme 1. These compounds were obtained by two steps,22,23 which are elucidated in the experimental section, and their structures are shown in Table 1. All of them are first reported and characterized by 1H NMR, elemental analysis, melting test and mass spectra, which were in full accordance with their depicted structures. The structure of S12 was also characterized by 13C NMR spectrum.All the synthesized aryl dihydrothiazol acyl podophyllotoxin ester derivatives S1–S17 were evaluated for their anti-proliferative activities against three lung cancer cell lines, A549, Calu-1, 973 and two normal cell lines, Vero (African green monkey kidney cells) and L02 (human normal liver cells) by MTT assay and the results are shown in Table 2. Compared with podophyllotoxin itself, the introduction of aryl dihydrothiazol moieties generally attenuated the cytotoxicity of podophyllotoxin against the normal cells. However, the lethality of podophyllotoxin derivatives to lung cancer cells was not reduced apparently. Conversely, most dihydrothiazol moieties even improved the anti-proliferative activities of the agents. As listed in Table 2, the anti-proliferative activities of S3 (IC50 = 0.56 μM), S7 (IC50 = 1.94 μM), S8 (IC50 = 0.93 μM), S12 (IC50 = 0.18 μM), S14 (IC50 = 0.79 μM) and S17 (IC50 = 0.95 μM) are better than podophyllotoxin (IC50 = 6.57 μM) itself and the positive control, Combretastatin A-4 (CA-4) (IC50 = 2.78 μM) against A549 cells. For Calu-1 cells, the anti-proliferative effects of S2 (IC50 = 2.34 μM), S7 (IC50 = 3.46 μM), S12 (IC50 = 3.03 μM), S14 (IC50 = 3.45 μM) and S15 (IC50 = 3.37 μM) are superior to podophyllotoxin (IC50 = 9.47 μM) and CA-4 (IC50 = 3.65 μM). In the case of 973 cells, S4 (IC50 = 3.12 μM), S7 (IC50 = 1.94 μM), S8 (IC50 = 2.95 μM), S11 (IC50 = 2.49 μM), S12 (IC50 = 1.63 μM) and S15 (IC50 = 3.54 μM) displayed good anti-proliferative properties. Through comparison, we found that S12 is the best agent, which should be selected for further study.
| Compound | Cytotoxicity (IC50a, μM) | ||||
|---|---|---|---|---|---|
| A549 | Calu-1 | 973 | L02 | Vero | |
| a Data are the mean of three independent experiments.b CA-4: Combretastatin A-4. | |||||
| S1 | 7.36 | 12.5 | 31.4 | >100 | >100 |
| S2 | 14.62 | 2.34 | 9.19 | >100 | >100 |
| S3 | 0.56 | 11.7 | 7.23 | >100 | >100 |
| S4 | 9.57 | 9.71 | 3.12 | >100 | >100 |
| S5 | 16.41 | 14.29 | 28.5 | >100 | >100 |
| S6 | 13.88 | 16.26 | 21.2 | >100 | >100 |
| S7 | 1.94 | 3.46 | 1.94 | >100 | >100 |
| S8 | 0.93 | 13.8 | 2.95 | >100 | >100 |
| S9 | 10.9 | 28.9 | 6.23 | >100 | >100 |
| S10 | 3.01 | 8.25 | 16.9 | >100 | >100 |
| S11 | 6.49 | 12.3 | 2.49 | >100 | >100 |
| S12 | 0.18 | 3.03 | 1.63 | >100 | >100 |
| S13 | 8.18 | 24.6 | 36.5 | >100 | >100 |
| S14 | 0.79 | 3.45 | 13.7 | >100 | >100 |
| S15 | 13.62 | 3.37 | 3.54 | >100 | >100 |
| S16 | 11.7 | 6.02 | 31.4 | >100 | >100 |
| S17 | 0.95 | 18.16 | 16.68 | >100 | >100 |
| Podophyllotoxin | 6.57 | 9.47 | 5.58 | 3.16 | 1.04 |
| CA-4 | 2.78 | 3.65 | 4.76 | 4.23 | 2.56 |
After that, we investigated the effect of S12 on cell apoptosis. We treated A549 cells with varying concentrations (0, 0.18, 0.37, 0.75 μM) of S12 for 24 h and analysed cells for changes in apoptotic markers by flow cytometer in vitro. As is shown in Fig. 2, after treatment with increasing concentrations of S12, the percentage of apoptotic cell increased slightly. In view of the fact that the increase is not very significant, we speculated that the apoptosis inducing effect of S12 is moderate.
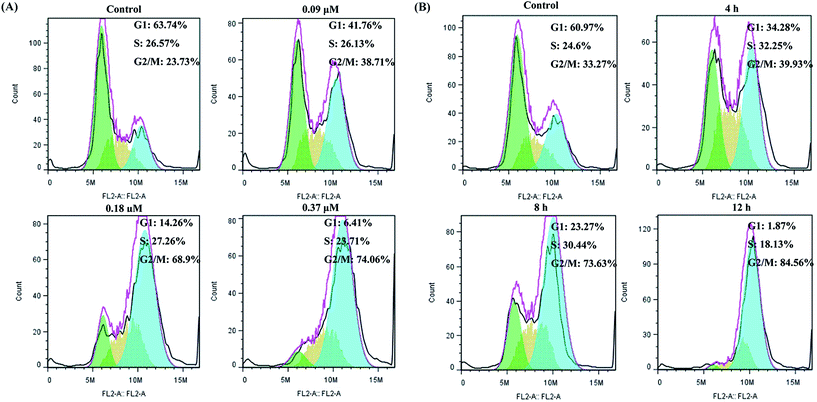
We next assessed the cell cycle distribution of A549 cells by flow cytometry. Treatment of A549 cells with S12 at various concentrations (0, 0.09, 0.18, 0.37 μM) for 8 h and treated cells with 0.18 μM S12 for different times (0, 4, 8, 12 h) could both arrest cells at G2/M phase. From Fig. 3(A), we can see that treatment of A549 cells with varying doses of S12 for 8 hours resulted in increased accumulation of the cells in G2/M phase. When the drug concentration rose to 0.37 μM, 74.06% of cells were arrested at G2/M phase. In a time-dependent experiment, maximum accumulation (84.56%) of cells in the G2/M phase was observed after treatment of cells with 0.18 μM S12 for 12 hours (Fig. 3(B)). Remarkably, S12 can cause a significant cell cycle arrest at G2/M phase at a low dose within a short time.
For better understanding of the potency of S12 and to guide further SAR studies, we examined the interaction of S12 with tubulin (PDB code: 1SA0). All docking runs applied the Lamarckian genetic algorithm of Auto-Dock 4.0. The interaction of S12 with tubulin amino acid residues is depicted in Fig. 4(A). All the amino acid residues of tubulin which had interactions with S12 were exhibited. In the binding model, S12 is nicely bound to the colchicine binding site of tubulin via one π bond with LYS 254 (distance = 4.56 Å) and two hydrogen bonds with CYS 241 (distance = 2.47 Å and 1.92 Å). In Fig. 4(B), 3D models of the interaction between S12 and tubulin are depicted. The molecular docking results argue that S12 may be a potential tubulin polymerization inhibitor.
 | ||
| Fig. 4 Molecular docking analysis of S12, showing proposed binding modes at the colchicine binding pocket β-tubulin (PDB code: 1SA0). Hydrogen bonds are displayed as lime dashed lines and π bond is displayed as an orange line. (A) Interaction of S12 with the amino acid residues of colchicine binding site (carbon atom, gray; oxygen atom, red; nitrogen atom, purple; sulphur atom, yellow, chlorine atom, green). (B) Binding position of S12 in the protein surface of tubulin (carbon atom, gray; oxygen atom, red; nitrogen atom, purple; sulphur atom, yellow, chlorine atom, green; hydrogen atom, white). | ||
In order to observe the phenotypic effect of S12 on the cellular cytoskeletal network of tubulin, A549 cells were immunostained and analysed under a confocal microscope. As illustrated in Fig. 5, substantial de-polymerization of microtubules is demonstrated in this assay. Compared with the control group, S12 treated cells induced substantial destabilization of microtubules and formed polymorphonuclear cells. The cellular morphology of S12 treated A549 is the same as or even more remarkable than the positive control group colchicine treated cells.
 | ||
| Fig. 5 Effect of S12 (0.5 μM) on interphase microtubules of A549 cells. Microtubules tagged with rhodamine (red) and nuclei tagged with DAPI (blue) were observed under a confocal microscope. | ||
To further investigate the effect of S12 on microtubule organization, we did an in vitro microtubule assembly assay. As shown in Fig. 6, S12 caused a decrease in the microtubule assembly (the curve shifts to the left when compared with the control group), which was the same as colchicine and in contrast to paclitaxel. To sum up, our results demonstrate that S12 inhibits tubulin polymerization, blocks mitosis, and causes cell death.
Experimental
General information and materials
All chemicals (reagent grade) used were purchased from Nanjing Chemical Reagent Co. Ltd (China). Melting points (uncorrected) were measured on a XT4 MP Apparatus (Taike Corp., Beijing, China). All 1H NMR and 13C NMR spectra were recorded on a Bruker DPX 300 or DRX 500 spectrometer in CDCl3. Chemical shifts (δ) for 1H NMR spectra were reported in ppm (δ). ESI mass spectra were obtained on a Mariner Biospectrometry Workstation (ESI-TOF) mass spectrometer. Elemental analyses were performed on a CHN-O-Rapid instrument and were within 0.4% of the theoretical values. TLC was carried out on the glass-backed silica gel sheets (silica gel 60 Å GF254) and visualized in UV light (254 nm).Goat anti-mouse IgG (H + L) was purchased from Invitrogen Trading (Shanghai) Co., Ltd (Shanghai, China). β-Tubulin antibody (#2146) was purchased from Cell Signaling Technology (Beverly, MA). Anti-tubulin (#AT819), Cy3-labeled goat anti-mouse IgG (H + L) (#A0521) were purchased from Cytoskeleton, Inc. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) were purchased from Beyotime Institute of Biotechnology (Haimen, China). RNase A (#EN0531) was purchased from Thermo Scientific, Fermentas (USA). AnnexinV-FITC cell apoptosis assay kit (#BA11100) was purchased from BIO-BOX (Nanjing, China).
General procedure for preparation of compounds a1–a17
A mixture of aryl nitrile (30 mmol), L-cysteine (60 mmol), and NaHCO3 (120 mmol) in EtOH (1.5 mL mmol−1) was stirred at 100 °C, until TLC analysis indicated disappearance of the nitrile. The solvent was evaporated under reduced pressure and H2O (5 mL mmol−1) was added to the residual white solid. The cold solution was then acidified with HCl to pH 2. The precipitated solid was collected by filtration and washed with a small amount of H2O.General procedure for preparation of compounds S1–S17
Aryl dihydrothiazol acids derivatives a1–a17, podophyllotoxin, 4-dimethyaminopyridine (DMAP) and N,N′-dicyclohexylcarbodiimide (DCC) were dissolved in dichloromethane and stirred for 12 h. A proper amount of silica gel was added and the solvent was condensed by vacuum concentration. Then, the target compounds were collected by column chromatography (V(acetone)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) V(dichloromethane) = 50
V(dichloromethane) = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Chemical structures of the target compounds (S1–S17) are shown in Table 1.
1). Chemical structures of the target compounds (S1–S17) are shown in Table 1.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.40 (t, J = 8.7 Hz, 1H, O
O), 5.40 (t, J = 8.7 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.61 (s, 1H, CH-Ar), 4.42 (dd, J = 9.1, 6.5 Hz, 1H, CH–CH2–O), 4.22 (t, J = 9.9 Hz, 1H, CH–CH2–O), 3.81 (s, 3H, 4′-OCH3), 3.74 (dd, J = 7.7, 2.3 Hz, 2H, S–CH2), 3.71 (s, 6H, 3′,5′-OCH3), 2.99–2.91 (m, 2H, O
C–CH–N), 4.61 (s, 1H, CH-Ar), 4.42 (dd, J = 9.1, 6.5 Hz, 1H, CH–CH2–O), 4.22 (t, J = 9.9 Hz, 1H, CH–CH2–O), 3.81 (s, 3H, 4′-OCH3), 3.74 (dd, J = 7.7, 2.3 Hz, 2H, S–CH2), 3.71 (s, 6H, 3′,5′-OCH3), 2.99–2.91 (m, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C32H29NO9S ([M + Na]+) 626.1563, found 626.1162. Anal. Calcd for C32H29NO9S: C, 63.67; H, 4.84; N, 2.32; O, 23.85; S, 5.31. Found: C, 62.33; H, 4.89; N, 2.56; O, 23.99; S, 5.16.
C–CH, O–CH2–CH). ESI-TOF, calcd for C32H29NO9S ([M + Na]+) 626.1563, found 626.1162. Anal. Calcd for C32H29NO9S: C, 63.67; H, 4.84; N, 2.32; O, 23.85; S, 5.31. Found: C, 62.33; H, 4.89; N, 2.56; O, 23.99; S, 5.16.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.40 (td, J = 8.8, 3.4 Hz, 1H, O
O), 5.40 (td, J = 8.8, 3.4 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.61 (s, 1H, CH-Ar), 4.42 (dt, J = 14.4, 7.3 Hz, 1H, CH–CH2–O), 4.24 (dd, J = 19.2, 9.1 Hz, 1H, CH–CH2–O), 3.81 (s, 3H, 4′-OCH3), 3.72 (d, J = 5.9 Hz, 6H, 3′,5′-OCH3), 3.73 (d, J = 10.3, 5.1 Hz, 2H, S–CH2), 2.93 (d, J = 14.8 Hz, 2H, O
C–CH–N), 4.61 (s, 1H, CH-Ar), 4.42 (dt, J = 14.4, 7.3 Hz, 1H, CH–CH2–O), 4.24 (dd, J = 19.2, 9.1 Hz, 1H, CH–CH2–O), 3.81 (s, 3H, 4′-OCH3), 3.72 (d, J = 5.9 Hz, 6H, 3′,5′-OCH3), 3.73 (d, J = 10.3, 5.1 Hz, 2H, S–CH2), 2.93 (d, J = 14.8 Hz, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH), 2.40 (d, J = 5.6 Hz, 3H, –CH3). ESI-TOF, calcd for C33H31NO9S ([M + Na]+) 640.1720, found 640.1562. Anal. Calcd for C33H31NO9S: C, 64.17; H, 5.06; N, 2.27; O, 23.31; S, 5.19. Found: C, 63.35; H, 5.29; N, 2.34; O, 23.46; S, 5.15.
C–CH, O–CH2–CH), 2.40 (d, J = 5.6 Hz, 3H, –CH3). ESI-TOF, calcd for C33H31NO9S ([M + Na]+) 640.1720, found 640.1562. Anal. Calcd for C33H31NO9S: C, 64.17; H, 5.06; N, 2.27; O, 23.31; S, 5.19. Found: C, 63.35; H, 5.29; N, 2.34; O, 23.46; S, 5.15.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.39 (t, J = 8.0 Hz, 1H, O
O), 5.39 (t, J = 8.0 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.61 (s, 1H, CH-Ar), 4.42 (d, J = 5.1 Hz, 1H, CH–CH2–O), 4.29–4.16 (m, 1H, CH–CH2–O), 3.81 (s, 3H, 4′-OCH3), 3.87–3.61 (m, 2H, S–CH2), 3.71 (d, J = 4.0 Hz, 6H, 3′,5′-OCH3), 2.95 (s, 2H, O
C–CH–N), 4.61 (s, 1H, CH-Ar), 4.42 (d, J = 5.1 Hz, 1H, CH–CH2–O), 4.29–4.16 (m, 1H, CH–CH2–O), 3.81 (s, 3H, 4′-OCH3), 3.87–3.61 (m, 2H, S–CH2), 3.71 (d, J = 4.0 Hz, 6H, 3′,5′-OCH3), 2.95 (s, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH), 2.40 (d, J = 4.0 Hz, 3H, –CH3). ESI-TOF, calcd for C33H31NO9S ([M + Na]+) 640.1720, found 640.1562. Anal. Calcd for C33H31NO9S: C, 64.17; H, 5.06; N, 2.27; O, 23.31; S, 5.19. Found: C, 63.35; H, 5.29; N, 2.34; O, 23.46; S, 5.15.
C–CH, O–CH2–CH), 2.40 (d, J = 4.0 Hz, 3H, –CH3). ESI-TOF, calcd for C33H31NO9S ([M + Na]+) 640.1720, found 640.1562. Anal. Calcd for C33H31NO9S: C, 64.17; H, 5.06; N, 2.27; O, 23.31; S, 5.19. Found: C, 63.35; H, 5.29; N, 2.34; O, 23.46; S, 5.15.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.39 (d, J = 8.2 Hz, 1H, O
O), 5.39 (d, J = 8.2 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.61 (s, 1H, CH-Ar), 4.42 (s, 1H, CH–CH2–O), 4.24 (d, J = 6.1 Hz, 1H, CH–CH2–O), 3.91–3.76 (m, 5H, S–CH2, 4′-OCH3), 3.71 (s, 6H, 3′,5′-OCH3), 2.95 (s, 2H, O
C–CH–N), 4.61 (s, 1H, CH-Ar), 4.42 (s, 1H, CH–CH2–O), 4.24 (d, J = 6.1 Hz, 1H, CH–CH2–O), 3.91–3.76 (m, 5H, S–CH2, 4′-OCH3), 3.71 (s, 6H, 3′,5′-OCH3), 2.95 (s, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C33H31NO10S ([M + Na]+) 656.1669, found 656.1325. Anal. Calcd for C33H31NO10S: C, 62.55; H, 4.93; N, 2.21; O, 25.25; S, 5.06. Found: C, 62.30; H, 5.01; N, 2.32; O, 25.33; S, 5.00.
C–CH, O–CH2–CH). ESI-TOF, calcd for C33H31NO10S ([M + Na]+) 656.1669, found 656.1325. Anal. Calcd for C33H31NO10S: C, 62.55; H, 4.93; N, 2.21; O, 25.25; S, 5.06. Found: C, 62.30; H, 5.01; N, 2.32; O, 25.33; S, 5.00.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.41 (t, J = 8.7 Hz, 1H, O
O), 5.41 (t, J = 8.7 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.63 (s, 1H, CH-Ar), 4.43 (dd, J = 7.6, 5.1 Hz, 1H, CH–CH2–O), 4.25 (dt, J = 15.4, 9.9 Hz, 1H, CH–CH2–O), 3.83 (s, 3H, 4′-OCH3), 3.78 (dd, J = 13.8, 5.3 Hz, 2H, S–CH2), 3.75 (s, 3H, –OCH3), 3.75–3.69 (s, 6H, 3′,5′-OCH3), 2.96 (s, 2H, O
C–CH–N), 4.63 (s, 1H, CH-Ar), 4.43 (dd, J = 7.6, 5.1 Hz, 1H, CH–CH2–O), 4.25 (dt, J = 15.4, 9.9 Hz, 1H, CH–CH2–O), 3.83 (s, 3H, 4′-OCH3), 3.78 (dd, J = 13.8, 5.3 Hz, 2H, S–CH2), 3.75 (s, 3H, –OCH3), 3.75–3.69 (s, 6H, 3′,5′-OCH3), 2.96 (s, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C33H31NO10S ([M + Na]+) 656.1669, found 656.1325. Anal. Calcd for C33H31NO10S: C, 62.55; H, 4.93; N, 2.21; O, 25.25; S, 5.06. Found: C, 62.30; H, 5.01; N, 2.32; O, 25.33; S, 5.00. ESI-TOF, calcd for C33H28F3NO10S ([M + Na]+) 694.1437, found 694.1089. Anal. Calcd for C33H28F3NO10S: C, 59.01; H, 4.20; F, 8.49; N, 2.09; O, 21.44; S, 4.77. Found: C, 58.72; H, 4.32; F, 8.47; N, 2.17; O, 21.59; S, 4.75.
C–CH, O–CH2–CH). ESI-TOF, calcd for C33H31NO10S ([M + Na]+) 656.1669, found 656.1325. Anal. Calcd for C33H31NO10S: C, 62.55; H, 4.93; N, 2.21; O, 25.25; S, 5.06. Found: C, 62.30; H, 5.01; N, 2.32; O, 25.33; S, 5.00. ESI-TOF, calcd for C33H28F3NO10S ([M + Na]+) 694.1437, found 694.1089. Anal. Calcd for C33H28F3NO10S: C, 59.01; H, 4.20; F, 8.49; N, 2.09; O, 21.44; S, 4.77. Found: C, 58.72; H, 4.32; F, 8.47; N, 2.17; O, 21.59; S, 4.75.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.42 (t, J = 8.9 Hz, 1H, O
O), 5.42 (t, J = 8.9 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.62 (s, 1H, CH-Ar), 4.42 (dd, J = 9.2, 6.4 Hz, 1H, CH–CH2–O), 4.28–4.16 (m, 1H, CH–CH2–O), 3.81 (s, 3H, 4′-OCH3), 3.80–3.75 (m, 2H, S–CH2), 3.73 (s, 6H, 3′,5′-OCH3), 3.01–2.90 (m, 2H, O
C–CH–N), 4.62 (s, 1H, CH-Ar), 4.42 (dd, J = 9.2, 6.4 Hz, 1H, CH–CH2–O), 4.28–4.16 (m, 1H, CH–CH2–O), 3.81 (s, 3H, 4′-OCH3), 3.80–3.75 (m, 2H, S–CH2), 3.73 (s, 6H, 3′,5′-OCH3), 3.01–2.90 (m, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C33H28F3NO9S ([M + Na]+) 694.1437, found 694.1089. Anal. Calcd for C33H28F3NO9S: C, 59.01; H, 4.20; F, 8.49; N, 2.09; O, 21.44; S, 4.77. Found: C, 58.72; H, 4.32; F, 8.47; N, 2.17; O, 21.59; S, 4.75.
C–CH, O–CH2–CH). ESI-TOF, calcd for C33H28F3NO9S ([M + Na]+) 694.1437, found 694.1089. Anal. Calcd for C33H28F3NO9S: C, 59.01; H, 4.20; F, 8.49; N, 2.09; O, 21.44; S, 4.77. Found: C, 58.72; H, 4.32; F, 8.47; N, 2.17; O, 21.59; S, 4.75.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.39 (tt, J = 25.9, 12.8 Hz, 1H, O
O), 5.39 (tt, J = 25.9, 12.8 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.62 (s, 1H, CH-Ar), 4.42 (dd, J = 9.2, 6.2 Hz, 1H, CH–CH2–O), 4.24 (dd, J = 19.7, 9.4 Hz, 1H, CH–CH2–O), 3.81 (s, 3H, 4′-OCH3), 3.80–3.75 (m, 2H, S–CH2), 3.74 (d, J = 3.6 Hz, 6H, 3′,5′-OCH3), 2.95 (d, J = 1.9 Hz, 2H, O
C–CH–N), 4.62 (s, 1H, CH-Ar), 4.42 (dd, J = 9.2, 6.2 Hz, 1H, CH–CH2–O), 4.24 (dd, J = 19.7, 9.4 Hz, 1H, CH–CH2–O), 3.81 (s, 3H, 4′-OCH3), 3.80–3.75 (m, 2H, S–CH2), 3.74 (d, J = 3.6 Hz, 6H, 3′,5′-OCH3), 2.95 (d, J = 1.9 Hz, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C33H28F3NO10S ([M + Na]+) 710.1386, found 710.1082. Anal. Calcd for C33H28F3NO10S: C, 57.64; H, 4.10; F, 8.29; N, 2.04; O, 23.27; S, 4.66. Found: C, 57.45; H, 4.43; F, 8.43; N, 2.47; O, 21.97; S, 4.74.
C–CH, O–CH2–CH). ESI-TOF, calcd for C33H28F3NO10S ([M + Na]+) 710.1386, found 710.1082. Anal. Calcd for C33H28F3NO10S: C, 57.64; H, 4.10; F, 8.29; N, 2.04; O, 23.27; S, 4.66. Found: C, 57.45; H, 4.43; F, 8.43; N, 2.47; O, 21.97; S, 4.74.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.38 (t, J = 8.6 Hz, 1H, O
O), 5.38 (t, J = 8.6 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.59 (s, 1H, CH-Ar), 4.41 (dd, J = 9.1, 6.3 Hz, 1H, CH–CH2–O), 4.22 (dd, J = 19.9, 9.8 Hz, 1H, CH–CH2–O), 3.80 (s, 3H, 4′-OCH3), 3.71 (s, 6H, 3′,5′-OCH3), 3.77–3.62 (m, 2H, S–CH2), 3.02–2.87 (m, 3H, O
C–CH–N), 4.59 (s, 1H, CH-Ar), 4.41 (dd, J = 9.1, 6.3 Hz, 1H, CH–CH2–O), 4.22 (dd, J = 19.9, 9.8 Hz, 1H, CH–CH2–O), 3.80 (s, 3H, 4′-OCH3), 3.71 (s, 6H, 3′,5′-OCH3), 3.77–3.62 (m, 2H, S–CH2), 3.02–2.87 (m, 3H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH, –CH–(CH3)2), 1.26 (s, 3H, –CH–(CH3)2), 1.24 (s, 3H, –CH–(CH3)2). ESI-TOF, calcd for C35H35NO9S ([M + Na]+) 668.2033, found 668.1937. Anal. Calcd for C35H35NO9S: C, 65.10; H, 5.46; N, 2.17; O, 22.30; S, 4.97. Found: C, 64, 25; H, 5.58; N, 2.33; O, 22.57; S, 4.86.
C–CH, O–CH2–CH, –CH–(CH3)2), 1.26 (s, 3H, –CH–(CH3)2), 1.24 (s, 3H, –CH–(CH3)2). ESI-TOF, calcd for C35H35NO9S ([M + Na]+) 668.2033, found 668.1937. Anal. Calcd for C35H35NO9S: C, 65.10; H, 5.46; N, 2.17; O, 22.30; S, 4.97. Found: C, 64, 25; H, 5.58; N, 2.33; O, 22.57; S, 4.86.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.38 (td, J = 9.0, 3.9 Hz, 1H, O
O), 5.38 (td, J = 9.0, 3.9 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.62 (d, J = 2.6 Hz, 1H, CH-Ar), 4.43 (dd, J = 9.6, 5.4 Hz, 1H, CH–CH2–O), 4.24 (dd, J = 19.9, 9.6 Hz, 1H, CH–i2–O), 3.81 (d, J = 3.0 Hz, 3H, 4′-OCH3), 3.73 (s, 6H, 3′,5′-OCH3), 3.77–3.65 (m, 2H, S–CH2), 3.00–2.88 (m, 2H, O
C–CH–N), 4.62 (d, J = 2.6 Hz, 1H, CH-Ar), 4.43 (dd, J = 9.6, 5.4 Hz, 1H, CH–CH2–O), 4.24 (dd, J = 19.9, 9.6 Hz, 1H, CH–i2–O), 3.81 (d, J = 3.0 Hz, 3H, 4′-OCH3), 3.73 (s, 6H, 3′,5′-OCH3), 3.77–3.65 (m, 2H, S–CH2), 3.00–2.88 (m, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C32H28FNO9S ([M + Na]+) 643.1469, found 643.1232. Anal. Calcd for C32H28FNO9S: C, 61.83; H, 4.54; F, 3.06; N, 2.25; O, 23.16; S, 5.16. Found: C, 61.01; H, 4.64; F, 3.02; N, 2.43; O, 23.27; S, 5.04.
C–CH, O–CH2–CH). ESI-TOF, calcd for C32H28FNO9S ([M + Na]+) 643.1469, found 643.1232. Anal. Calcd for C32H28FNO9S: C, 61.83; H, 4.54; F, 3.06; N, 2.25; O, 23.16; S, 5.16. Found: C, 61.01; H, 4.64; F, 3.02; N, 2.43; O, 23.27; S, 5.04.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.52–5.43 (m, 1H, O
O), 5.52–5.43 (m, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.63 (s, 1H, CH-Ar), 4.45 (d, J = 6.2 Hz, 1H, CH–CH2–O), 4.22 (d, J = 9.1 Hz, 1H, CH–CH2–O), 3.82 (s, 3H, –OCH3), 3.78 (s, 3H, 4′-OCH3), 3.71 (s, 6H, 3′,5′-OCH3), 3.49 (m, 2H, S–CH2), 2.97 (s, 2H, O
C–CH–N), 4.63 (s, 1H, CH-Ar), 4.45 (d, J = 6.2 Hz, 1H, CH–CH2–O), 4.22 (d, J = 9.1 Hz, 1H, CH–CH2–O), 3.82 (s, 3H, –OCH3), 3.78 (s, 3H, 4′-OCH3), 3.71 (s, 6H, 3′,5′-OCH3), 3.49 (m, 2H, S–CH2), 2.97 (s, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C33H30FNO10S ([M + Na]+) 674.1574, found 674.1346. Anal. Calcd for C33H30FNO10S: C, 60.82; H, 4.64; F, 2.92; N, 2.15; O, 24.55; S, 4.92. Found: C, 60.08; H, 4.92; F, 3.00; N, 2.33; O, 24.77; S, 4.84.
C–CH, O–CH2–CH). ESI-TOF, calcd for C33H30FNO10S ([M + Na]+) 674.1574, found 674.1346. Anal. Calcd for C33H30FNO10S: C, 60.82; H, 4.64; F, 2.92; N, 2.15; O, 24.55; S, 4.92. Found: C, 60.08; H, 4.92; F, 3.00; N, 2.33; O, 24.77; S, 4.84.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.40 (t, J = 8.6 Hz, 1H, O
O), 5.40 (t, J = 8.6 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.61 (s, 1H, CH-Ar), 4.40 (s, 1H, CH–CH2–O), 4.22 (s, 1H, CH–CH2–O), 3.77 (d, J = 15.2 Hz, 5H, 4′-OCH3, S–CH2), 3.71 (s, 6H, 3′,5′-OCH3), 2.95 (s, 2H, O
C–CH–N), 4.61 (s, 1H, CH-Ar), 4.40 (s, 1H, CH–CH2–O), 4.22 (s, 1H, CH–CH2–O), 3.77 (d, J = 15.2 Hz, 5H, 4′-OCH3, S–CH2), 3.71 (s, 6H, 3′,5′-OCH3), 2.95 (s, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C32H27ClFNO9S ([M + Na]+) 678.1079, found 678.1002. Anal. Calcd for C32H27ClFNO9S: C, 58.58; H, 4.15; Cl, 5.40; F, 2.90; N, 2.13; O, 21.95; S, 4.89. Found: C, 58.03; H, 4.22; Cl, 5.49; F, 3.05; N, 2.36; O, 22.41; S, 4.72.
C–CH, O–CH2–CH). ESI-TOF, calcd for C32H27ClFNO9S ([M + Na]+) 678.1079, found 678.1002. Anal. Calcd for C32H27ClFNO9S: C, 58.58; H, 4.15; Cl, 5.40; F, 2.90; N, 2.13; O, 21.95; S, 4.89. Found: C, 58.03; H, 4.22; Cl, 5.49; F, 3.05; N, 2.36; O, 22.41; S, 4.72.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.38 (t, J = 8.7 Hz, 1H, O
O), 5.38 (t, J = 8.7 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.61 (s, 1H, CH-Ar), 4.48–4.36 (m, 1H, CH–CH2–O), 4.29–4.14 (m, 1H, CH–CH2–O), 3.82 (s, 3H, 4′-OCH3), 3.78–3.74 (m, 2H, S–CH2), 3.73 (s, 6H, 3′,5′-OCH3), 3.01–2.86 (m, 2H, O
C–CH–N), 4.61 (s, 1H, CH-Ar), 4.48–4.36 (m, 1H, CH–CH2–O), 4.29–4.14 (m, 1H, CH–CH2–O), 3.82 (s, 3H, 4′-OCH3), 3.78–3.74 (m, 2H, S–CH2), 3.73 (s, 6H, 3′,5′-OCH3), 3.01–2.86 (m, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). 13C NMR (CDCl3, 300 MHz): δ 173.34 (13 C), 171.16 (14 C), 170.29 (17 C), 152.56 (3′,5′-C), 148.23 (7 C), 147.6 (6 C), 138.05 (4′-C), 137.07 (4′′-C), 134.52 (1′-C), 132.45 (1′′-C), 130.78 (9 C), 129.74 (2′′, 6′′-C), 128.79 (3′′, 5′′-C), 127.56 (10 C), 109.71 (8 C), 107.96 (2′, 6′-C), 106.79 (5 C), 101.58 (13 C), 78.24 (15 C), 75.02 (4 C), 71.13 (11 C), 60.66 (4′-OCH3), 56.01 (3′,5′-OCH3), 45.43 (1 C), 43.63 (2 C), 38.43 (3 C), 33.84 (16 C). ESI-TOF, calcd for C32H28ClNO9S ([M + Na]+) 660.1173, found 660.1084. Anal. Calcd for C32H28ClNO9S: C, 60.23; H, 4.42; Cl, 5.56; N, 2.20; O, 22.57; S, 5.03. Found: C, 59.79; H, 4.63; Cl, 5.54; N, 2.29; O, 22.74; S, 5.00.
C–CH, O–CH2–CH). 13C NMR (CDCl3, 300 MHz): δ 173.34 (13 C), 171.16 (14 C), 170.29 (17 C), 152.56 (3′,5′-C), 148.23 (7 C), 147.6 (6 C), 138.05 (4′-C), 137.07 (4′′-C), 134.52 (1′-C), 132.45 (1′′-C), 130.78 (9 C), 129.74 (2′′, 6′′-C), 128.79 (3′′, 5′′-C), 127.56 (10 C), 109.71 (8 C), 107.96 (2′, 6′-C), 106.79 (5 C), 101.58 (13 C), 78.24 (15 C), 75.02 (4 C), 71.13 (11 C), 60.66 (4′-OCH3), 56.01 (3′,5′-OCH3), 45.43 (1 C), 43.63 (2 C), 38.43 (3 C), 33.84 (16 C). ESI-TOF, calcd for C32H28ClNO9S ([M + Na]+) 660.1173, found 660.1084. Anal. Calcd for C32H28ClNO9S: C, 60.23; H, 4.42; Cl, 5.56; N, 2.20; O, 22.57; S, 5.03. Found: C, 59.79; H, 4.63; Cl, 5.54; N, 2.29; O, 22.74; S, 5.00.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.49–5.36 (m, 1H, O
O), 5.49–5.36 (m, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.62 (s, 1H, CH-Ar), 4.49–4.37 (m, 1H, CH–CH2–O), 4.31–4.16 (m, 1H, CH–CH2–O), 3.88–3.74 (m, 2H, S–CH2), 3.80 (s, 3H, 4′-OCH3), 3.71 (s, 6H, 3′,5′-OCH3), 2.97 (d, J = 9.7 Hz, 2H, O
C–CH–N), 4.62 (s, 1H, CH-Ar), 4.49–4.37 (m, 1H, CH–CH2–O), 4.31–4.16 (m, 1H, CH–CH2–O), 3.88–3.74 (m, 2H, S–CH2), 3.80 (s, 3H, 4′-OCH3), 3.71 (s, 6H, 3′,5′-OCH3), 2.97 (d, J = 9.7 Hz, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C32H27Cl2NO9S ([M + Na]+) 694.0784, found 694.0039. Anal. Calcd for C32H27Cl2NO9S: C, 57.15; H, 4.05; Cl, 10.54; N, 2.08; O, 21.41 S, 4.77. Found: C, 58.92; H, 4.23; Cl, 10.64; N, 2.19; O, 21.75; S, 4.52.
C–CH, O–CH2–CH). ESI-TOF, calcd for C32H27Cl2NO9S ([M + Na]+) 694.0784, found 694.0039. Anal. Calcd for C32H27Cl2NO9S: C, 57.15; H, 4.05; Cl, 10.54; N, 2.08; O, 21.41 S, 4.77. Found: C, 58.92; H, 4.23; Cl, 10.64; N, 2.19; O, 21.75; S, 4.52.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.38 (t, J = 8.6 Hz, 1H, O
O), 5.38 (t, J = 8.6 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.61 (s, 1H, CH-Ar), 4.48–4.32 (m, 1H, CH–CH2–O), 4.28–4.17 (m, 1H, CH–i2–O), 3.81 (s, 3H, 4′-OCH3), 3.77 (dd, J = 8.9, 4.4 Hz, 2H, S–CH2), 3.72 (s, 6H, 3′,5′-OCH3), 3.05–2.87 (m, 2H, O
C–CH–N), 4.61 (s, 1H, CH-Ar), 4.48–4.32 (m, 1H, CH–CH2–O), 4.28–4.17 (m, 1H, CH–i2–O), 3.81 (s, 3H, 4′-OCH3), 3.77 (dd, J = 8.9, 4.4 Hz, 2H, S–CH2), 3.72 (s, 6H, 3′,5′-OCH3), 3.05–2.87 (m, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C32H28BrNO9S ([M + Na]+) 704.0668, found 704.0274. Anal. Calcd for C32H28BrNO9S: C, 56.31; H, 4.13; Br, 11.71; N, 2.05; O, 21.10 S, 4.70. Found: C, 55.82; H, 4.43; Br, 11.68; N, 2.15; O, 21.19 S, 4.64.
C–CH, O–CH2–CH). ESI-TOF, calcd for C32H28BrNO9S ([M + Na]+) 704.0668, found 704.0274. Anal. Calcd for C32H28BrNO9S: C, 56.31; H, 4.13; Br, 11.71; N, 2.05; O, 21.10 S, 4.70. Found: C, 55.82; H, 4.43; Br, 11.68; N, 2.15; O, 21.19 S, 4.64.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.37 (td, J = 8.8, 3.9 Hz, 1H, O
O), 5.37 (td, J = 8.8, 3.9 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.60 (s, 1H, CH-Ar), 4.45–4.33 (m, 1H, CH–CH2–O), 4.22 (dd, J = 19.5, 9.7 Hz, 1H, CH–CH2–O), 3.80 (s, 3H, 4′-OCH3), 3.72 (d, J = 4.3 Hz, 6H, 3′,5′-OCH3), 3.48 (t, J = 10.3 Hz, 2H, S–CH2), 2.91 (d, J = 11.6 Hz, 2H, O
C–CH–N), 4.60 (s, 1H, CH-Ar), 4.45–4.33 (m, 1H, CH–CH2–O), 4.22 (dd, J = 19.5, 9.7 Hz, 1H, CH–CH2–O), 3.80 (s, 3H, 4′-OCH3), 3.72 (d, J = 4.3 Hz, 6H, 3′,5′-OCH3), 3.48 (t, J = 10.3 Hz, 2H, S–CH2), 2.91 (d, J = 11.6 Hz, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C32H28INO9S ([M + Na]+) 752.0529, found 752.0341. Anal. Calcd for C32H28INO9S: C, 56.68; H, 3.87; I, 17.40; N, 1.92; O, 19.71 S, 4.40. Found: C, 56.12; H, 3.97; I, 17.42; N, 2.02; O, 19.97 S, 4.41.
C–CH, O–CH2–CH). ESI-TOF, calcd for C32H28INO9S ([M + Na]+) 752.0529, found 752.0341. Anal. Calcd for C32H28INO9S: C, 56.68; H, 3.87; I, 17.40; N, 1.92; O, 19.71 S, 4.40. Found: C, 56.12; H, 3.97; I, 17.42; N, 2.02; O, 19.97 S, 4.41.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.40 (t, J = 8.7 Hz, 1H, O
O), 5.40 (t, J = 8.7 Hz, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.61 (s, 1H, CH-Ar), 4.42 (dd, J = 9.0, 6.2 Hz, 1H, CH–CH2–O), 4.23 (dd, J = 16.5, 9.5 Hz, 1H, CH–CH2–O), 3.86–3.73 (m, 5H, 4′-OCH3, S–CH2), 3.71 (s, 6H, 3′,5′-OCH3), 2.92 (d, J = 14.7 Hz, 2H, O
C–CH–N), 4.61 (s, 1H, CH-Ar), 4.42 (dd, J = 9.0, 6.2 Hz, 1H, CH–CH2–O), 4.23 (dd, J = 16.5, 9.5 Hz, 1H, CH–CH2–O), 3.86–3.73 (m, 5H, 4′-OCH3, S–CH2), 3.71 (s, 6H, 3′,5′-OCH3), 2.92 (d, J = 14.7 Hz, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C32H27Cl2NO9S ([M + Na]+) 694.0784, found 694.0039. Anal. Calcd for C32H27Cl2NO9S: C, 57.15; H, 4.05; Cl, 10.54; N, 2.08; O, 21.41 S, 4.77. Found: C, 58.92; H, 4.23; Cl, 10.64; N, 2.19; O, 21.75; S, 4.52.
C–CH, O–CH2–CH). ESI-TOF, calcd for C32H27Cl2NO9S ([M + Na]+) 694.0784, found 694.0039. Anal. Calcd for C32H27Cl2NO9S: C, 57.15; H, 4.05; Cl, 10.54; N, 2.08; O, 21.41 S, 4.77. Found: C, 58.92; H, 4.23; Cl, 10.64; N, 2.19; O, 21.75; S, 4.52.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 5.44–5.34 (m, 1H, O
O), 5.44–5.34 (m, 1H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH–N), 4.61 (s, 1H, CH-Ar), 4.48–4.38 (m, 1H, CH–CH2–O), 4.26 (t, J = 9.6 Hz, 1H, CH–CH2–O), 3.94 (dd, J = 6.0, 2.5 Hz, 6H, –OCH3), 3.81 (d, J = 2.2 Hz, 3H, 4′-OCH3), 3.72 (d, J = 12.0 Hz, 6H, 3′,5′-OCH3), 3.48 (ddd, J = 10.4, 9.2, 3.9 Hz, 2H, S–CH2), 2.95 (m, 2H, O
C–CH–N), 4.61 (s, 1H, CH-Ar), 4.48–4.38 (m, 1H, CH–CH2–O), 4.26 (t, J = 9.6 Hz, 1H, CH–CH2–O), 3.94 (dd, J = 6.0, 2.5 Hz, 6H, –OCH3), 3.81 (d, J = 2.2 Hz, 3H, 4′-OCH3), 3.72 (d, J = 12.0 Hz, 6H, 3′,5′-OCH3), 3.48 (ddd, J = 10.4, 9.2, 3.9 Hz, 2H, S–CH2), 2.95 (m, 2H, O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH, O–CH2–CH). ESI-TOF, calcd for C34H33NO11S ([M + Na]+) 686.1774, found 684.1243. Anal. Calcd for C34H33NO11S: C, 61.53; H, 5.01; N, 2.11; O, 26.52 S, 4.83. Found: C, 60.92; H, 5.23; N, 2.29; O, 26.75; S, 4.72.
C–CH, O–CH2–CH). ESI-TOF, calcd for C34H33NO11S ([M + Na]+) 686.1774, found 684.1243. Anal. Calcd for C34H33NO11S: C, 61.53; H, 5.01; N, 2.11; O, 26.52 S, 4.83. Found: C, 60.92; H, 5.23; N, 2.29; O, 26.75; S, 4.72.Cell lines and culture conditions
A549, Calu-1, 973, Vero and L02 cells were purchased from Nanjing Keygen Technology (Nanjing, China). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone) (High Glucose) with L-glutamine supplemented with 10% fetal bovine serum (FBS, BI), 100 U mL−1 penicillin and 100 μg mL−1 streptomycin (Hyclone), and incubated at 37 °C in a humidified atmosphere containing 5% CO2.Anti-proliferation assay
Target tumor cell lines were grown to log phase in DMEM medium supplemented with 10% fetal bovine serum. After diluting to 2 × 104 cells per mL with the complete medium, 100 μL of the obtained cell suspension was added to each well of 96-well culture plates and then allowed to adhere for 12 hours at 37 °C, 5% CO2 atmosphere. Tested samples at pre-set concentrations (0.1, 1, 10, 100 μM) were added to 96 wells with podophyllotoxin and CA-4 as positive references.After 24 hours exposure period, 20 μL PBS containing 2.5 mg mL−1 MTT was added to each well. Plates were then incubated for a further 4 hours, and then were centrifuged (1500 rpm at 4 °C for 10 minutes) to remove the supernatant. 150 μL DMSO was added to each well for coloration. The plates were shaken vigorously to ensure complete solubilization for 10 minutes at room temperature. The absorbance was measured and recorded on an ELISA reader (ELx800, BioTek, USA) at a test wavelength of 570 nm. In all experiments three replicate wells were used for each drug concentration. Each assay was carried out at least three times. The results are summarized in Table 2.
Cell apoptosis assay
For Annexin V/PI assays, A549 cells were stained with Annexin V-FITC and PI and then monitored for apoptosis by flow cytometry. Briefly, 5 × 103 cells were seeded in 6-well plates for 24 hours and then were treated with S12 (0, 0.18, 0.37, 0.75 μM) for 24 hours. Then cells were collected and washed twice with PBS and stained with 5 μL Annexin V-FITC and 2.5 μL PI (5 μg mL−1) in 1× binding buffer (10 mM HEPES, pH 7.4, 140 mM NaOH, 2.5 mM CaCl2) for 15 minutes at room temperature in the dark. Apoptotic cells were quantified using a BD Accuri C6 Flow Cytometer (BD, USA). Statistical analysis was done using Flowjo 7.6.1 software. Both early apoptotic (AnnexinV-positive, PI-negative) and late apoptotic (double positive of Annexin V and PI) cells were detected.Cell cycle analysis
A549 cells were plated in 6-well plates (5.0 × 103 cells per well) and incubated at 37 °C for 24 hours. Exponentially growing cells were then incubated with S12 at 0.09, 0.18 and 0.37 μM for 8 h. In the time-dependent assays, exponentially growing cells were incubated with 0.18 μM S12 at 37 °C for 4, 8 and 12 h. After that, cells were centrifuged and fixed in 70% ethanol at 4 °C for at least 12 hours and subsequently resuspended in PBS containing 0.1 mg mL−1 RNase A and 5 μg mL−1 propidium iodide (PI). Cellular DNA content, for cell cycle distribution analysis, was measured by flow cytometry using a BD Accuri C6 Flow Cytometer (BD, USA) plotting 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events per sample. The percentage of cells in the G1, S and G2/M phases of the cell cycle were determined using Flowjo 7.6.1 software after cell debris exclusion.
000 events per sample. The percentage of cells in the G1, S and G2/M phases of the cell cycle were determined using Flowjo 7.6.1 software after cell debris exclusion.
Docking simulation
The three-dimensional X-ray structure of tubulin (PDB code: 1SA0) was chosen as the template for the modelling study of S12 bound to tubulin. The crystal structure was obtained from the RCSB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). The molecular docking procedure was performed using Ligand Fit protocol within Auto-Dock 4.0. For ligand preparation, the 3D structure of S12 was generated and minimized using Auto-Dock 4.0. For protein preparation, hydrogen atoms were added, and the water and impurities were removed. The whole tubulin was defined as a receptor and the site sphere was selected based on the ligand binding location of taxol, then the taxol molecule was removed and S12 was placed during the molecular docking procedure. The types of interactions of the docked protein with ligand were analysed after the end of molecular docking.Confocal microscopy assay
A549 cells were grown on round cover slips to 70% confluence and incubated with 0.5 μM S12 and 2 μM colchicine for 12 hours, respectively. After incubating, cells were washed with PBS three times and fixed with 4% paraformaldehyde for 20 minutes, permeabilized with 1% TritonX-100 for another 10 minutes. Then, the cells were blocked with 3% BSA for 1 hour. Subsequently, the cells were washed once with PBS, and incubated with anti-tubulin antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Cytoskeleton, Inc.) in 3% BSA overnight at 4 °C. After being washed with 0.5% TritonX-100 (incubate for 5 minutes), each coverslip was added to 200 μL Cy3-labeled goat anti-mouse IgG (H + L) (1
500, Cytoskeleton, Inc.) in 3% BSA overnight at 4 °C. After being washed with 0.5% TritonX-100 (incubate for 5 minutes), each coverslip was added to 200 μL Cy3-labeled goat anti-mouse IgG (H + L) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1500, Cytoskeleton, Inc.) in 3% BSA and incubated for 1 hour at room temperature followed by DAPI (5 ng mL−1). Cells were then observed under an Olympus confocal microscope and data was analysed using FV-10-ASW 1.7 viewer.
1500, Cytoskeleton, Inc.) in 3% BSA and incubated for 1 hour at room temperature followed by DAPI (5 ng mL−1). Cells were then observed under an Olympus confocal microscope and data was analysed using FV-10-ASW 1.7 viewer.
Flow cytometry analysis of the expression of extracellular polymerized tubulin
A549 cells were seeded in 6-well plates and treated with 0.5 μM S12, 1 μM colchicine and 1 μM paclitaxel for 24 hours. Then, cells were harvested, washed once with PBS and blocked with 1% BSA for 30 min on ice, then were centrifuged (2800 rpm at 4 °C for 5 minutes) to remove supernatant. After that, cells were incubated with β-tubulin antibody for 1 h on ice and were centrifuged (2800 rpm at 4 °C for 5 minutes) to remove supernatant. Next, cells were incubated with secondary antibody (IgG) on ice for 1 h and were also centrifuged (2800 rpm at 4 °C for 5 minutes) to remove supernatant. Finally, cells were then resuspended in PBS and the expression level of polymerized tubulin was analysed by flow cytometry (BD, USA).Conclusion
In this paper, a series of novel aryl dihydrothiazol acyl podophyllotoxin ester derivatives S1–S17 were obtained through a two-step semi-synthetic route. Their anticancer biological activities were also evaluated. Among them, S12 showed the best anti-proliferative effect against A549 cell line with an IC50 value of 0.18 μM, which is better than podophyllotoxin (IC50 = 6.57 μM) and CA-4 (IC50 = 2.78 μM), and showed lower cytotoxicity against normal cells. Cell cycle distribution assay and apoptosis assay demonstrated that S12 can cause a remarkable cell cycle arrest at G2/M phase but the effect on apoptosis is not very significant. Furthermore, the docking simulation result enabled us to determine that tubulin is a target for S12. Confocal microscopy assay and extracellular polymerized tubulin expression analysis further confirmed that S12 really displayed its’ anticancer effect by inhibiting tubulin polymerization. To sum up, these findings prompt us to consider it as a potent anti-cancer agent.Abbreviations
| CA-4 | Combretastatin A-4 |
| NMR | Nuclear magnetic resonance spectrum |
| TLC | Thin layer chromatography |
| TMS | Tetramethylsilane |
| DMSO | Dimethyl sulfoxide |
| PBS | Phosphate-buffered saline |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| BSA | Bovine serum albumin |
Acknowledgements
The authors are grateful to the Program for Changjiang Scholars and Innovative Research Team in University (IRT_14R27).Notes and references
- D. M. Parkin, F. Bray, J. Ferlay and P. Pisani, Ca-Cancer J. Clin., 2002, 55, 74–108 CrossRef.
- E. C. Bruin, N. Granahan, R. Mitter, M. Salm, D. C. Wedge, L. Yates, M. J. Hanjani, S. Shafi, N. Murugaesu, A. J. Rowan, E. Grönroos, M. A. Muhammad, S. Horswell, M. Gerlinger, I. Varela, D. Jones, J. Marshall, T. Voet, P. V. Loo, D. M. Rassl, R. C. Rintoul, S. M. Janes, S. M. Lee, M. Forster, T. Ahmad, D. Lawrence, M. Falzon, A. Capitanio, T. T. Harkins, C. C. Lee, W. Tom, E. Teefe, S. C. Chen, S. Begum, A. Rabinowitz, B. Phillimore, B. S. Dene, G. Stamp, Z. Szallasi, N. Matthews, A. Stewart, P. Campbell and C. Swanton, Science, 2014, 346, 251–256 CrossRef PubMed.
- S. Sun, J. H. Schiller and A. F. Gazdar, Nat. Rev. Cancer, 2007, 7, 779–790 CrossRef PubMed.
- J. Wen, J. H. Fu, W. Zhang and M. Guo, Mod. Pathol., 2011, 24, 932–943 CrossRef PubMed.
- R. Siegel, D. Naishadham and A. Jemal, Ca-Cancer J. Clin., 2012, 62, 10–29 CrossRef PubMed.
- G. P. Pfeifer, M. F. Denissenko, M. Olivier, N. Tretyakova, S. S. Hecht and P. Hainaut, Oncogene, 2002, 21, 7435–7451 CrossRef CAS PubMed.
- J. Zhou, Z. Qu, S. Yan, F. Sun, J. A. Whitsett, S. D. Shapiro and G. Xiao, Oncogene, 2014, 1–11 Search PubMed.
- A. H. W. Williams, X. D. Dai, W. Blot, Z. Y. Xu, X. W. Sun, H. P. Xiao, B. J. Stone, S. F. Yu, Y. P. Feng, A. G. Ershow, J. Sun, J. F. Fraumeni and B. E. Henderson, Br. J. Cancer, 1990, 62, 982–987 CrossRef.
- O. Barbora, L. Noemie, P. Jana, D. Mario and D. Marc, Cancer Treat. Res., 2014, 159, 123–143 Search PubMed.
- R. Kaur, K. Kapoor and H. Kaur, J. Nat. Prod. Plant Resour., 2011, 1, 119–124 Search PubMed.
- A. Kamal, J. R. Tamboli, V. L. Nayak, S. F. Adil, M. V. P. S. Vishnuvardhan and S. Ramakrishna, Bioorg. Med. Chem., 2014, 22, 2714–2723 CrossRef CAS PubMed.
- Y. L. Qi, F. Liao, C. Q. Zhao, Y. D. Lin and M. X. Zuo, Acta Pharmacol. Sin., 2005, 26, 1000–1008 CrossRef CAS PubMed.
- A. Kamal, B. A. Kuma, P. Suresh, A. Juvekar and S. Zingde, Bioorg. Med. Chem., 2011, 19, 2975–2979 CrossRef CAS PubMed.
- S. H. Abdullah, S. Zhao, G. S. Mittal and O. D. Baik, Sep. Purif. Technol., 2012, 93, 92–97 CrossRef CAS PubMed.
- W. Zhao, L. Chen, H. M. Li, D. J. Wang, D. S. Li, T. Chen, Z. P. Yuan and Y. J. Tang, Bioorg. Med. Chem., 2014, 22, 2998–3007 CrossRef CAS PubMed.
- L. Xiao, W. Zhao, H. M. Li, D. J. Wan, D. S. Li, T. Chen and Y. J. Tang, Eur. J. Med. Chem., 2014, 80, 267–277 CrossRef CAS PubMed.
- J. K. Bai, W. Zhao, H. M. Li and Y. J. Tang, Curr. Med. Chem., 2012, 19, 927–936 CrossRef CAS.
- K. H. Lee, S. A. Beers, M. Mori, Z. Wang, Y. H. Kuo, L. Li, S. Y. Liu, J. Y. Chang, F. S. Han and Y. C. Chen, J. Med. Chem., 1990, 33, 1364–1368 CrossRef CAS.
- Z. Q. Wang, Y. H. Kuo, D. Schnur, J. P. Bowen, S. Y. Liu, F. S. Han, Y. C. Cheng and K. H. Lee, J. Med. Chem., 1990, 33, 2660–2666 CrossRef CAS.
- S. Marjan, A. Mohsen, N. O. Seyed, H. R. Gholam, A. Amir, S. Bentolhoda and S. Abbas, Bioorg. Med. Chem., 2013, 21, 7648–7654 CrossRef PubMed.
- D. M. Wang, J. W. Kuo, W. T. Kuo, C. F. Hsu, M. H. Wang, Y. Chang, W. J. Lin and J. T. Wang, J. Labelled Compd. Radiopharm., 2014, 57, 132–135 CrossRef CAS PubMed.
- O. V. Maltsev, V. Walter, M. J. Brandl and L. Hintermann, Synthesis, 2013, 45, 2763–2767 CrossRef CAS PubMed.
- H. Y. Lin, W. Chen, J. Shi, W. Y. Kong, J. L. Qi, X. M. Wang and Y. H. Yang, Chem. Biol. Drug Des., 2013, 81, 275–283 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra01871d |
| This journal is © The Royal Society of Chemistry 2015 |