Encapsulation of doxorubicin within multifunctional gadolinium-loaded dendrimer nanocomplexes for targeted theranostics of cancer cells†
Jingyi Zhua,
Zhijuan Xiongb,
Mingwu Shenb and
Xiangyang Shi*ab
aState Key Laboratory for Modification of Chemical Fibers and Polymer Materials, College of Materials Science and Engineering, Donghua University, Shanghai 201620, People's Republic of China
bCollege of Chemistry, Chemical Engineering and Biotechnology, Donghua University, Shanghai 201620, People's Republic of China. E-mail: xshi@dhu.edu.cn
First published on 23rd March 2015
Abstract
We report the use of multifunctional gadolinium-loaded dendrimer nanocomplexes to encapsulate an anticancer drug doxorubicin (DOX) for magnetic resonance (MR) imaging and chemotherapy of cancer cells. In this study, amine-terminated generation five poly(amidoamine) dendrimers (G5.NH2) were modified with chelator/gadolinium (Gd) complexes and folic acid (FA) via a polyethylene glycol (PEG) spacer, followed by acetylation of the remaining dendrimer terminal amines. The thus formed G5.NHAc–DOTA(Gd)–PEG–FA complexes were used to encapsulate DOX within the dendrimer interior. We show that the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes having 8.5 DOX molecules and DOTA/Gd complexes per dendrimer are stable under different pH conditions, and are able to release DOX in a sustained manner. The FA modification enables efficient targeting of the particles to cancer cells overexpressing FA receptors (FAR), and thus effective targeted MR imaging of the cancer cells in vitro. Likewise, the encapsulation of DOX within the dendrimer/Gd complexes does not compromise the therapeutic efficacy of the DOX drug. Importantly, by virtue of the FA-directed targeting, the formed multifunctional dendrimeric nanocomplexes are able to exert specific therapeutic efficacy of DOX to the FAR-overexpressing cancer cells in vitro. The developed multifunctional dendrimers with both MR imaging agents Gd(III) complexed via the conjugated chelator and anticancer drug physically encapsulated within the dendrimer interior may hold great promise to be used as a theranostic nanoplatform for targeted MR imaging and chemotherapy of different types of cancer.
Introduction
Recent advances in nanotechnology enable the development of various nanoparticulate systems incorporating both diagnosis and therapy functionalities for theranostics of cancer.1–8 This is because nanoparticles (NPs) are able to be designed to have targeting, imaging, and therapeutic functionalities,9–15 to have extended blood circulation half-life,16,17 and to overcome the drawbacks of conventional diagnostic and therapeutic agents, such as chelator/Gd(III) complexes used for magnetic resonance (MR) imaging18 or anticancer drugs19 (short blood circulation time and nonspecificity). The key to develop an effective theranostic nanomedicine for imaging-guided cancer therapy is to incorporate both imaging and therapeutic elements within a single nanoparticulate platform.20,21Theranostic NPs can be readily fabricated since the synthetic procedure is typically a combination of well-established chemistries originally developed solely for imaging or therapy. On one hand, the developed multifunctional NPs can be used as both contrast agents for molecular imaging applications and therapeutic agents under certain conditions in the absence of anticancer drugs. For instance, hyaluronic acid-modified Fe3O4@Au core/shell nanostars can be simultaneously used for MR/computed tomography (CT)/thermal tri-mode imaging and photothermal therapy of tumors.22 In another work, Zhou et al. demonstrated the use of tungsten oxide nanorods as an efficient nanoplatform for tumor CT imaging and photothermal therapy.23 On the other hand, NPs can be incorporated with both imaging agents for molecular imaging applications and anticancer drugs for chemotherapy applications. For instance, Kim et al. developed a drug-loaded aptamer–Au NP bioconjugate that could be used for combined CT imaging and therapy of prostate cancer.3 In another work,6 peptide-functionalized multifunctional polymeric micelles loaded with doxorubicin (DOX) and a cluster of superparamagnetic iron oxide NPs inside the micelle core can be used for targeted MR imaging and chemotherapy of cancer cells.
For theranostics of cancer via molecular imaging and chemotherapy, it is essential to find a versatile nanoplatform that can be used for efficient incorporation of the contrast agents and anticancer drugs. Among the many used nanoplatforms, dendrimers, especially poly(amidoamine) (PAMAM) dendrimers have attracted a great deal of interest.24–26 Dendrimers possess high monodispersity, branched internal cavity, and abundant surface functional groups.27 These properties enable dendrimers to be covalently modified with targeting agents, imaging molecules, and anticancer drugs for targeted drug delivery applications.25,28–32 Likewise, the relatively hydrophobic interior of the dendrimers allows for effective encapsulation of hydrophobic anticancer drugs.25,33–37 Furthermore, the unique structural characteristics of dendrimers afford their uses as templates or stabilizers to form dendrimer-entrapped gold NPs (Au DENPs)38,39 or dendrimer stabilized gold NPs40,41 for CT imaging applications, and as a platform to covalently modify chelator/Gd complexes for MR imaging applications.42,43 Even for low generation dendrimers (e.g., generation 2, G2), the PEGylation conjugation of the dendrimer periphery also enables the formation of Au DENPs for CT imaging applications.44,45 These advantages of PAMAM dendrimers render their uses as an ideal multifunctional nanoplatform for both imaging and drug delivery applications. However, there are few reports related to the simultaneous incorporation of both imaging elements and anticancer drugs into a single dendrimer-based nanoplatform for cancer theranostic applications.46–48
Inspired by our previous successes in the development of dendrimer-based MR imaging contrast agents42,43 and dendrimer-based drug delivery systems,25,34,35 we attempted to develop a dendrimer-based theranostic platform for targeted MR imaging and chemotherapy of cancer cells. In this study, amine-terminated G5 PAMAM dendrimers (G5.NH2) were modified with gadolinium (Gd) chelator and targeting ligand folic acid (FA) via a PEG spacer, followed by Gd(III) chelation and acetylation of the remaining dendrimer terminal amines. The formed mutifunctional dendrimers (G5.NHAc–DOTA(Gd)–PEG–FA) were used to physically encapsulate DOX. The final G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes were characterized via different techniques. The release kinetics, targeting specificity to FA receptors (FAR)-overexpressing cancer cells, therapeutic activity, and targeted therapeutic efficacy to cancer cells were investigated in vitro. Likewise, the T1-relaxometry of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes and the subsequent targeted MR imaging of cancer cells in vitro using the complexes were also assessed. To our knowledge, this is the first report demonstrating the potential to use dendrimers as a versatile nanoplatform to link chelator/Gd complexes on the periphery and encapsulate anticancer drug within the dendrimer interior for theranostics of cancer cells.
Experimental
Materials
Ethylenediamine core G5.NH2 PAMAM dendrimers with a polydispersity index less than 1.08 were purchased from Dendritech (Midland, MI). 2,2′,2′′-(10-(2-(2,5-Dioxopyrrolidin-1-yloxy)-2-oxoethyl)-1,4,7,10-tetraazacyclododecane-1,4,7-triy)triacetic acid (DOTA–NHS) was purchased from CheMatech (Dijon, France). PEG with one end of amine group and the other end of carboxyl group (NH2–PEG–COOH, Mw = 2000) and PEG monomethyl ether with the other end of carboxyl group (mPEG–COOH, Mw = 2000) were purchased from Shanghai Yanyi Biotechnology Corporation (Shanghai, China). DOX hydrochloride (DOX·HCl) was purchased from Beijing Huafeng Pharmaceutical Co., Ltd. (Beijing, China). Unless otherwise stated, the term of “free DOX” represents DOX·HCl. FA, acetic anhydride, triethylamine, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and all the other chemicals and solvents were purchased from Aldrich (St. Louis, MO) and used as received. KB cells (a human epithelial carcinoma cell line) were obtained from Institute of Biochemistry and Cell Biology (the Chinese Academy of Sciences, Shanghai, China). RPMI 1640 medium, fetal bovine serum (FBS), penicillin, and streptomycin were purchased from Hangzhou Jinuo Biomedical Technology (Hangzhou, China). Water used in all experiments was purified using a Milli-Q Plus 185 water purification system (Millipore, Bedford, MA) with resistivity higher than 18 MΩ cm. Regenerated cellulose dialysis membranes (molecular weight cut-off (MWCO) = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 or 1000) were acquired from Fisher (Pittsburgh, PA).
000 or 1000) were acquired from Fisher (Pittsburgh, PA).
Synthesis of the G5.NHAc–DOTA(Gd)–PEG–FA dendrimers
FA (5.14 mg, 0.0117 mmol) with about 1.5 molar equivalents of NH2–PEG–COOH (15.54 mg, 0.00777 mmol) dissolved in 5 mL DMSO was mixed with a DMSO solution (2 mL) containing EDC (6.75 mg, 0.0352 mmol), and the reaction mixture was stirred for 3 h to activated the γ-carboxylic acid group of FA. Then the activated FA was dropwise added to a DMSO solution (5 mL) containing NH2–PEG–COOH (46.93 mg, 0.0235 mmol) under vigorous magnetic stirring at room temperature. The reaction was stopped after 24 h. Then, the reaction mixture was extensively dialyzed against phosphate buffered saline (PBS, 3 times, 4 L) and water (3 times, 4 L) through a 1000 MWCO membrane for 3 days to remove the excess of reactants, followed by lyophilization to get the product FA–PEG–COOH.G5.NH2 (20.0 mg) dissolved in DMSO (5 mL) was reacted with 10 molar equivalents of DOTA–NHS (5.86 mg, 5 mL in DMSO) under vigorous magnetic stirring. The reaction was stopped after 24 h to get the raw product of G5.NH2–DOTA. In a parallel experiment, FA–PEG–COOH (34.6 mg) dissolved in 5.0 mL DMSO with 20 molar equivalents of G5.NH2 was reacted with EDC (29.5 mg, in 5.0 mL DMSO) under vigorous magnetic stirring for 3 h. The activated FA–PEG–COOH was then dropwise added into the DMSO solution of the raw product of the G5.NH2–DOTA solution under vigorous magnetic stirring. The reaction was continued for 3 days to obtain the raw product of G5.NH2–DOTA–PEG–FA conjugate. Then, Gd(NO3)3 (10.4 mg) dissolved in 1 mL water (with Gd(NO3)3/DOTA molar ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added under vigorous stirring to chelate Gd(III) ions. After 30 min, triethylamine (71.35 μL) was added to the reaction mixture solution and the solution was thoroughly mixed for 30 min. Excess acetic anhydride (40.38 μL, 5 molar equivalents of the dendrimer terminal amines) was then dropwise added into the solution under vigorous magnetic stirring. The mixture was allowed to react at room temperature with stirring for 24 h. Then the reaction mixture was purified and lyophilized to get the G5.NHAc–DOTA(Gd)–PEG–FA dendrimer according to the procedure similar to the purification of FA–PEG–COOH, except that a dialysis membrane with MWCO of 14
1) was added under vigorous stirring to chelate Gd(III) ions. After 30 min, triethylamine (71.35 μL) was added to the reaction mixture solution and the solution was thoroughly mixed for 30 min. Excess acetic anhydride (40.38 μL, 5 molar equivalents of the dendrimer terminal amines) was then dropwise added into the solution under vigorous magnetic stirring. The mixture was allowed to react at room temperature with stirring for 24 h. Then the reaction mixture was purified and lyophilized to get the G5.NHAc–DOTA(Gd)–PEG–FA dendrimer according to the procedure similar to the purification of FA–PEG–COOH, except that a dialysis membrane with MWCO of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 was used. For comparison, non-targeted G5.NHAc–DOTA(Gd)–mPEG dendrimer was also synthesized and characterized under similar experimental conditions except that mPEG was used to conjugate the G5 dendrimer.
000 was used. For comparison, non-targeted G5.NHAc–DOTA(Gd)–mPEG dendrimer was also synthesized and characterized under similar experimental conditions except that mPEG was used to conjugate the G5 dendrimer.
Encapsulation of DOX within the G5.NHAc–DOTA(Gd)–PEG–FA dendrimers
G5.NHAc–DOTA(Gd)–PEG–FA dendrimers (21.3 mg) were dissolved in 1.5 mL water. DOX·HCl with 10 molar equivalents of dendrimers was dissolved in 300 μL methanol and was neutralized with 5 μL triethylamine to generate deprotonated DOX, which is insoluble in water. Then the DOX solution was mixed with the 1.5 mL dendrimer aqueous solution. The mixture solution was vigorously stirred overnight to allow the evaporation of the methanol solvent. The dendrimer–DOX mixture solution was centrifuged (7000 rpm for 10 min) to remove the precipitates related to the non-complexed deprotonated DOX. The precipitate was collected and dissolved into 1 mL methanol for UV-vis spectroscopic analysis. The supernatant was lyophilized to obtain the dendrimer/DOX complex.Characterization techniques
1H NMR spectra were recorded using Bruker AV-400 NMR spectrometer. All samples were dissolved in D2O before measurements. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry was performed using a 4800 Plus MALDI TOF/TOF™ Analyzer (AB SCIEX, Framingham, MA). Linear mode was selected as the operation mode. Beta-indoleacrylic acid (10 mg mL−1) in acetonitrile/H2O (v/v = 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) was used as the matrix and 1 mg of the dendrimer sample was dissolved in 1 mL methanol. The dendrimer solution was well mixed with an equal volume of the matrix solution. Then, 1 μL solution of the mixture was injected on the spots of the target plate before measurements. UV-vis spectra were collected using a Lambda 25 UV-vis spectrophotometer (Perkin Elmer, Waltham, MA). Samples were dissolved in water before measurements. Zeta-potential measurements were carried out using a Malvern Zetasizer Nano ZS system (Worcestershire, UK) equipped with a standard 633 nm laser. Dendrimer samples with a concentration of 1 mg mL−1 were measured under different pH conditions (pH 5.0, 7.4 and 10.0).
30) was used as the matrix and 1 mg of the dendrimer sample was dissolved in 1 mL methanol. The dendrimer solution was well mixed with an equal volume of the matrix solution. Then, 1 μL solution of the mixture was injected on the spots of the target plate before measurements. UV-vis spectra were collected using a Lambda 25 UV-vis spectrophotometer (Perkin Elmer, Waltham, MA). Samples were dissolved in water before measurements. Zeta-potential measurements were carried out using a Malvern Zetasizer Nano ZS system (Worcestershire, UK) equipped with a standard 633 nm laser. Dendrimer samples with a concentration of 1 mg mL−1 were measured under different pH conditions (pH 5.0, 7.4 and 10.0).
In vitro release kinetic study
G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes dissolved in water (1 mg, 1 mL) were separately placed in a dialysis bag with an MWCO of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, hermetically tied, and suspended in 9 mL of aqueous medium (PBS (pH 7.4) or acetate buffer (pH 5.0)). The entire system was kept in a vapor-bathing constant temperature vibrator at 37 °C. One milliliter of the buffer medium was taken out at each predetermined time interval and measured by UV-vis spectrometry. The volume of the outer phase buffer medium was maintained constant by replenishing 1 mL of the corresponding buffer solution. For comparison, DOX·HCl dissolved in water (1 mg, 1 mL) was also subjected to a similar release experiment using PBS (pH 7.4) as a releasing medium.
000, hermetically tied, and suspended in 9 mL of aqueous medium (PBS (pH 7.4) or acetate buffer (pH 5.0)). The entire system was kept in a vapor-bathing constant temperature vibrator at 37 °C. One milliliter of the buffer medium was taken out at each predetermined time interval and measured by UV-vis spectrometry. The volume of the outer phase buffer medium was maintained constant by replenishing 1 mL of the corresponding buffer solution. For comparison, DOX·HCl dissolved in water (1 mg, 1 mL) was also subjected to a similar release experiment using PBS (pH 7.4) as a releasing medium.
Cell culture
KB cells were regularly cultured and passaged in RPMI 1640 medium supplemented with 10% FBS, 100 U mL−1 penicillin, and 100 U mL−1 streptomycin at 37 °C in a 5% CO2 incubator. The KB cells grown in FA-free medium express high-level FAR (denoted as KB-HFAR), while the cells grown in FA-containing medium (2.5 μM FA) for 24 h or longer express low-level FAR (denoted as KB-LFAR).Cytotoxicity assay and cell morphology observation
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was used to check the therapeutic efficacy of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes in vitro. Briefly, 1 × 104 KB-HFAR cells per well were seeded into a 96-well plate. After overnight incubation to bring the cells to confluence, the medium was replaced with 200 μL fresh medium containing free DOX, G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes with the same DOX concentration, and G5.NHAc–DOTA(Gd)–PEG–FA dendrimers with the same dendrimer concentration of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes pre-dissolved in 20 μL PBS. After 24 h incubation at 37 °C, an MTT solution (20 μL in PBS, 5 mg mL−1) was added to each well and the cells were incubated for another 4 h at 37 °C. After that, the medium in each well was carefully removed, and DMSO (200 μL) was added to dissolve the formed formazan crystals. The assays were carried out according to the manufacturer's instructions at an absorption wavelength of 570 nm using a Thermo Scientific Multiskan MK3 ELISA reader (Thermo Scientific, Waltham, MA). For each sample, mean and standard deviation for the triplicate wells were analyzed.After treatment with free DOX (600 nM), G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes with the same DOX concentration, and G5.NHAc–DOTA(Gd)–PEG–FA dendrimers with the same dendrimer concentration of G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes for 24 h according to the above conditions, the morphology of cells was observed using a Leica DM IL LED inverted phase contrast microscope.
Flow cytometry analysis
Flow cytometry was used to check the targeting specificity of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes to KB-HFAR cells by monitoring the DOX fluorescence. Both KB-HFAR and KB-LFAR cells were seeded in 12-well cell culture plates at a density of 2 × 105 cells per well the day before the experiment, then the cells were rinsed 4 times with serum-free FA-deficient RPMI 1640 medium and cultured with fresh FA-deficient RPMI 1640 medium containing free DOX (5 μM), G5.NHAc–DOTA(Gd)–mPEG/DOX complexes, and G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes with a DOX concentration of 5 μM. After 3 h incubation, the cells were trypsinized, resuspended in PBS containing 0.1% bovine serum albumin, and analyzed using a Becton Dickinson FACScan analyzer equipped with a 15 mW, 488 nm, and air-cooled argon ion laser. The fluorescent emission was collected through a 575 nm band-pass filter and acquired in log mode. The FL2-fluorescence of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells was measured, and the mean fluorescence of gated viable cells was quantified.
000 cells was measured, and the mean fluorescence of gated viable cells was quantified.
Confocal microscopy
Confocal microscopy (Carl Zeiss LSM 700, Jena, Germany) was used to observe the specific uptake of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes by KB-HFAR cells according to our previous reports.49,50 Briefly, cover slips with a diameter of 14 mm were pretreated with 5% HCl, 30% HNO3, and 75% alcohol and then fixed in a 24-well tissue culture plate. 8 × 104 KB cells were seeded into each well with 1 mL fresh medium and cultured at 37 °C and 5% CO2 for 24 h to allow the cells to attach onto the cover slips. Then, the medium was discarded, and replaced with 1 mL fresh medium containing PBS (control), free DOX (5 μM), G5.NHAc–DOTA(Gd)–mPEG/DOX complexes ([DOX] = 5 μM), and G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes ([DOX] = 5 μM), respectively. After 3 h incubation at 37 °C and 5% CO2, the cells were rinsed with PBS for 3 times, fixed with glutaraldehyde (2.5%) for 15 min at 4 °C, and counterstained with Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 (1 μg mL−1) for 20 min at 37 °C using a standard procedure. The emission was collected through a 505–525 nm barrier filter. The optical section thickness was set at 5 mm. Samples were scanned using a 63× oil-immersion objective lens.
342 (1 μg mL−1) for 20 min at 37 °C using a standard procedure. The emission was collected through a 505–525 nm barrier filter. The optical section thickness was set at 5 mm. Samples were scanned using a 63× oil-immersion objective lens.
Targeted inhibition of cancer cells in vitro
To evaluate the efficiency of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes for targeted cancer cell inhibition, MTT assay was performed. Briefly, 1 × 104 KB-HFAR or KB-LFAR cells per well were seeded into a 96-well plate. After overnight incubation to bring the cells to confluence, the medium was replaced with fresh medium containing G5.NHAc–DOTA(Gd)–mPEG/DOX or G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes at the DOX concentration of 1 μM. The cells were incubated at 37 °C and 5% CO2 for 3 h, rinsed with PBS for 3 times, and cultured with fresh regular DOX-free medium for another 24 h before MTT assay. KB-HFAR cells treated with PBS were used as control. The MTT assay was performed as described above.T1 MR relaxometry
T1 MR relaxometry of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes was performed using a 0.5 T NMI20-Analyst NMR Analyzing and Imaging system (Shanghai NIUMAG Corporation, Shanghai, China). The samples were diluted in water with Gd(III) concentrations in the range of 0.05–0.8 mM. A total of four echoes were used with the following parameters: TR = 300 ms, TE = 18.2 ms, matrix = 192 × 256, section thickness = 3.8 mm, and FOV = 10 cm. MR imaging of each sample at different concentrations was performed. The T1 relaxivity (r1) was determined by a linear fitting of the inverse relaxation time (1/T1) as a function of Gd(III) concentration.Targeted MR imaging of cancer cells in vitro
KB-HFAR and KB-LFAR cells were separately seeded into a 6-well plate at a density of 3 × 106 cells per well and incubated in regular RPMI 1640 medium for 24 h. Then the medium was replaced with fresh medium containing the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes ([Gd] = 0, 0.5, 1, 2, and 4 μM, respectively). After incubation at 37 °C and 5% CO2 for 3 h, the cells were washed 3 times with PBS, trypsinized, centrifuged, and resuspended in 1 mL PBS (containing 0.5% agarose) in a 2 mL Eppendorf tube before MR imaging. T1 MR imaging of the cell suspension in each sample tube was performed by 0.5 T NMI20-Analyst NMR Analyzing and Imaging system using a wrist receiver coil with a SPIN ECHO sequence (TR = 700 ms, TE = 12 ms, point resolution = 256 mm × 196 mm, section thickness = 5 mm, and FOV = 100 × 100 mm).In vitro cellular uptake assay
Both KB-HFAR and KB-LFAR cells were separately seeded in a 12-well plate at a density of 5 × 105 cells per well and incubated in regular RPMI 1640 medium for 24 h. Then, the medium was replaced with fresh medium containing the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes ([Gd] = 1 and 4 μM, respectively) and the cells were incubated at 37 °C and 5% CO2. After 3 h, the cells were washed 3 times with PBS, lifted with trypsinization, and resuspended in PBS. The cell suspensions (100 μL) were counted. The remaining cells were centrifuged to form pellets and lysed using an aqua regia solution (0.5 mL) to digest the cells. The digested cell sample was then diluted with 1.5 mL PBS before quantification by Leeman Prodigy inductively coupled plasma-optical emission spectroscopy (ICP-OES, Hudson, NH).Statistical analysis
One way ANOVA statistical analysis was performed to evaluate the significance of the experimental data. 0.05 was selected as the significance level, and the data were indicated with (*) for p < 0.05, (**) for p < 0.01, and (***) for p < 0.001, respectively.Results and discussion
Synthesis and characterization of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes
In the dendrimer-based drug delivery system, partial PEGylation of G5 PAMAM dendrimer enables improved drug loading efficiency within the dendrimer interior.51–53 Likewise, the modification of targeting ligand onto the G5 dendrimer surface via a PEG spacer enables enhanced targeting specificity of the dendrimer nanodevices.54 Therefore, similar to our previous studies,48,51,54 we modified FA onto the G5 dendrimer surface via a PEG spacer after the G5 dendrimer was modified with DOTA ligands. Followed by Gd(III) chelation and acetylation of the remaining dendrimer terminal amines, the formed multifunctional dendrimers were used to encapsulate an anticancer drug DOX (Fig. 1). | ||
| Fig. 1 Schematic illustration of the structure (a) and synthesis (b) of the designed dendrimeric nanoplatform for theranostics of cancer cells. | ||
The conjugation of DOTA ligands onto the surface of G5.NH2 dendrimers was characterized via 1H NMR spectroscopy. Because the –CH2– proton signals of DOTA overlap with those of the G5 dendrimer, we used an indirect method reported in our previous studies42,43 to calculate the number of DOTA ligands attached to each G5 dendrimer by NMR integration. Meanwhile, we also used MALDI-TOF mass spectrometry to calculate the actual number of DOTA ligands attached to each dendrimer by comparison of the Mw difference between the G5.NH2 and G5.NH2–DOTA dendrimers. 1H NMR spectroscopy results show that the number of DOTA moieties attached to each dendrimer is 8.9 (Fig. S1, ESI†), similar to our previous studies.42,43 The MALDI-TOF mass spectra demonstrate that around 8.5 DOTA molecules are attached on the surface of each dendrimer (Fig. S2, ESI†). Then, PEGylated FA formed by reacting FA with NH2–PEG–COOH (having 0.6 FA moieties linked to each PEG demonstrated via 1H NMR spectroscopy, Fig. S3, ESI†) was conjugated with the amines of G5 dendrimers. By comparison of the NMR integration, the number of PEG–FA conjugated onto each G5 dendrimer was estimated to be 19.4 (Fig. S4, ESI†). The control G5.NH2–DOTA–mPEG dendrimers without FA conjugation were also characterized using the same method and the attached numbers of DOTA and mPEG moieties onto each G5 dendrimer were comparable to those of the final G5.NH2–DOTA–PEG–FA dendrimers (Fig. S5, ESI†). Knowing the practical number of primary amines per G5 dendrimer,55 one can easily calculate the number of the remaining primary amines after each step reaction via NMR or MALDI-TOF.
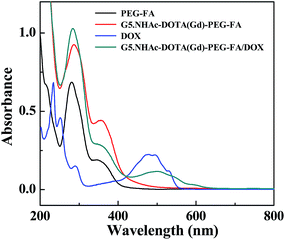
The formed G5.NH2–DOTA–PEG–FA dendrimers were then used to chelate Gd(III) ion, followed by acetylation of the remaining dendrimer terminal amines. The acetylation step rendered the dendrimers with a slightly positive surface potential (11.3 ± 2.3 mV, Table S1, ESI†), in agreement with the literature.38 The obtained G5.NHAc–DOTA(Gd)–PEG–FA dendrimers were utilized as a nanoplatform to physically encapsulate DOX. The formation of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes was characterized via UV-vis spectrometry (Fig. 2). Clearly, the apparent absorption peak at around 281 nm is associated to the FA absorption (similar to the PEG–FA segments and the G5.NHAc–DOTA(Gd)–PEG–FA dendrimers), and the peak at 491 nm is due to the characteristic absorption of DOX, suggesting the successful conjugation of FA and encapsulation of DOX. The slight red shift of DOX absorption peak for dendrimer/DOX complex when compared with that of free DOX might be due to the fact that the intermolecular interaction of DOX becomes weakened after being encapsulated within the dendrimers, hence leading to decreased electronic transition. The DOX payload within the G5.NHAc–DOTA(Gd)–PEG–FA dendrimers was analyzed with UV-vis spectroscopy using a standard DOX absorbance/concentration calibration curve. We show that there are 8.5 DOX molecules complexed with each G5.NHAc–DOTA(Gd)–PEG–FA dendrimer. The drug encapsulation efficiency (EE%) and drug loading percentage (DL%) were calculated to be 78.9% and 5.7%, respectively (Table S2, ESI†). It should be noted that the encapsulation efficiency of DOX within the G5.NHAc–DOTA(Gd)–PEG–FA dendrimer is greatly improved compared to that within the G5.NHAc–FI–FA dendrimer without PEGylation. This should be due to the fact that partial PEGylation of dendrimers is able to enlarge the dendrimer periphery, enabling enhanced drug loading, in agreement with our previous work.54 ICP-OES was used to analyze the loading of Gd(III) ions within the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes. We show that there are about 8.9 Gd(III) ions loaded within each G5 dendrimer through the DOTA chelation process, approximately similar to the number of DOTA ligands conjugated onto each G5 dendrimer.
The formed G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes are quite stable. We show that the lyophilized powder of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes are able to be dispersed well in aqueous solution, and are stable under different pH conditions (pH = 5.0, 7.4, and 10.0) (Fig. S6, ESI†). Zeta potential of the complexes was measured under different pH conditions (Table S1†). The surface potential of the dendrimers and complexes at pH 7.4 was measured to be 11.3 ± 2.3 and 8.8 ± 4.0 mV, respectively. It seems that the DOX loading within the dendrimers does not significantly alter the surface potential of the complexes. At an acidic pH condition (pH = 5.0), the surface potentials of both dendrimers and complexes are more positive than at the physiological condition (pH = 7.4). This is presumably due to the fact that at pH 5.0 protonation of a portion of dendrimer tertiary amines occurs.56 The surface potential change for both the dendrimers and the complexes with pH is similar to that for the G5.NHAc dendrimers described in our previous work.33
In vitro release kinetics
To exert the therapeutic efficacy, it is important to investigate the in vitro release kinetics of DOX from the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes. The cumulative release of DOX from the complexes shows that the drug is able to be released in a sustained manner (Fig. 3). In contrast, free DOX is quickly released to the outer phase of the dialysis tube in PBS, and about 69% of DOX is released within just 5 h, in agreement with the literature.34,51,57 In both buffers with different pH conditions, the release of DOX from the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes followed a pattern which is characterized by an initial faster release followed by a sustained release. In PBS (pH = 7.4), about 8.6% of the DOX drug is released within 1 h from the complexes, and about 26.9% of the DOX is released within 48 h. The drug release rate in the acetate buffer (pH 5.0) is slightly faster than that in PBS (pH 7.4). About 9.6% of DOX is released within 1 h and approximately 29.4% of the drug is released within 48 h. This should be due to the fact that under acidic condition, DOX is more protonated and more hydrophilic, enabling slightly fast release of DOX from the relatively hydrophobic dendrimer interior. Overall, the prolonged release profile of DOX from the complexes under both pH conditions implies that the relatively hydrophobic interior of the dendrimer molecules is extremely useful for effective encapsulation and retention of the hydrophobic DOX drug.Targeting specificity of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes
FAR has been known to be overexpressed by a range of human carcinomas, including breast, ovary, endometrium, kidney, lung, head and neck, brain, and myeloid cancers.58,59 Through the conjugation of FA onto the surface of dendrimers, the formed G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes are expected to have targeting specificity to cancer cells overexpressing FAR. The specific cellular uptake of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes was examined by flow cytometric analysis of KB cells (Fig. 4 and S7, ESI†) treated with the particles by virtue of the DOX fluorescence. It can be clearly seen that after 3 h incubation with the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes, KB-HFAR cells display significant fluorescence enhancement than KB-LFAR cells (Fig. 4a, S7e and f†). In contrast, both KB-HFARs and KB-LFARs cells treated with the G5.NHAc–DOTA(Gd)–mPEG/DOX complexes without FA do not display significantly enhanced fluorescence intensity (Fig. 4a, S7c and d†), but display slightly higher fluorescence intensity than the KB-HFAR cells treated with PBS (Fig. S7a†). This indicates that the binding of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes with KB-HFAR cells is quite specific due to the receptor-mediated targeting pathway, in agreement with the literature.60,61 The slightly higher fluorescence intensity of KB-HFAR cells treated with the G5.NHAc–DOTA(Gd)–mPEG/DOX complexes and KB-LFAR cells treated with either the G5.NHAc–DOTA(Gd)–mPEG/DOX or G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes may be ascribed to the non-specific cellular uptake of the particles through mechanisms of phagocytosis and diffusion via cell walls, in agreement with the literature.38,39 The cellular uptake of free DOX and the dendrimer/DOX complexes was also quantified by measuring the mean fluorescence intensity of cells and the population of cells uptaken with the complexes (Fig. 4a and b). The results are similar to the fluorescence histogram analysis data. Our results confirmed the binding specificity of the developed G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes to FAR-overexpressing cancer cells via a receptor-mediated manner.The fluorescence of DOX also enabled confocal microscopic imaging of the cellular uptake of the complexes (Fig. 5). It can be seen that after 3 h incubation with the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes, only KB-HFAR cells display significant red fluorescence signals, which are associated with the specific binding and internalization of the complexes into the cytoplasm of the cells. In contrast, under similar microscopic conditions, the KB-LFAR cells treated with the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes and both KB-LFAR and KB-HFAR cells treated with the G5.NHAc–DOTA(Gd)–mPEG/DOX complexes do not exhibit appreciable fluorescence signals. This result suggests that only the FA-modified dendrimeric complexes are able to be specifically uptaken by the FAR-overexpressing KB-HFAR cells. It is worth noting that similar to other nanoscale drug delivery system,34,62,63 the designed dendrimer/DOX complex is able to be trapped inside sub-cellular organelles, such as lysosome or endosome, and nuclei (due to the pores on the nuclei membrane) after 3 h exposure. Taken together with the flow cytometric data, our results clearly suggest that the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes can be specifically delivered to FAR-overexpressing cancer cells via receptor-mediated binding and endocytosis.
 | ||
| Fig. 5 Confocal microscopy images of KB-HFAR and KB-LFAR cells treated with different materials at the DOX concentration of 5 μM for 3 h, respectively. | ||
Therapeutic efficacy of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes in vitro
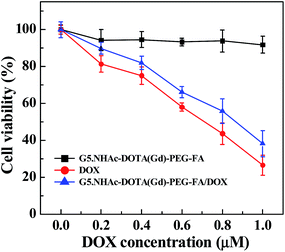
We next checked the therapeutic activity of the developed G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes via MTT cell viability assay. As shown in Fig. 6, the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes are able to inhibit the growth of cancer cells with the half-maximal inhibitory concentration (IC50) of 0.85 μM, which is slightly higher than that of free DOX (0.7 μM). The antitumor efficacy of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes is solely related to the loaded drug DOX, since the drug-free G5.NHAc–DOTA(Gd)–PEG–FA dendrimers do not display apparent cytotoxicity at the dendrimer concentrations equivalent to the studied DOX concentration range.The therapeutic efficacy of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes was further confirmed by microscopic visualization of the cell morphology change after treatment with the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes (Fig. S8, ESI†). It can be seen that the treatment of both free DOX (600 nM) and the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes with the same DOX concentration causes a significant cytotoxicity to KB cells, and a significant portion of cells became rounded and non-adherent, indicative of the cell death (Fig. S8d and e†). In contrast, no rounded and detached cells can be visualized in control cells without treatment (Fig. S8a†), cells treated with equivalent volume of PBS (Fig. S8b†), cells treated with DOX-free G5.NHAc–DOTA(Gd)–PEG–FA dendrimers (Fig. S8c†) with the dendrimer concentration similar to that of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes ([DOX] = 600 nM). Our results further suggest that the bioactivity of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes is solely associated with the loaded DOX drug. The cell morphology observation data corroborate the MTT assay results.
To prove our hypothesis that the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes are able to specifically inhibit the growth of KB-HFAR cells via FA-mediated targeting, after 3 h incubation of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes, the medium in wells was replaced with fresh medium. MTT assay was carried out after the cells were incubated for additional 24 h (Fig. 7). It is clear that only the treatment of KB-HFAR cells with the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes results in a significant decrease of the cell viability (65%). In contrast, the viability of KB-HFAR cells treated with the non-targeted G5.NHAc–DOTA(Gd)–mPEG/DOX complexes, and KB-LFAR cells treated with either the G5.NHAc–DOTA(Gd)–PEG–FA/DOX or the G5.NHAc–DOTA(Gd)–mPEG/DOX complexes is still around 80%, much higher than that of the KB-HFAR cells treated with the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes (p < 0.001). These results clearly indicate that the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes afford targeted inhibition of the growth of cancer cells via a receptor-mediated manner.
T1 MR relaxometry
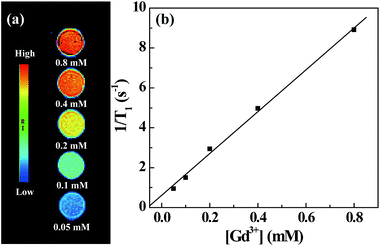
The presence of Gd(III) ions rendered the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes with r1 relaxivity for MR imaging applications. T1-weighted MR images show that the MR signal intensity of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes increases with the Gd concentration of the complexes (Fig. 8a). By plotting the inverse T1 (1/T1) as a function of the Gd concentration, a linear curve was obtained with a slope of 10.58 mM−1 s−1 (Fig. 8b), which can be defined to be the longitudinal relaxivity (r1) of the complexes. It should be noted that the r1 relaxivity of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes is much higher than that of the G5-Gd dendrimers (4.84 mM−1 s−1) reported in our previous work.43 This could be due to the fact that the interior DOX encapsulation may lead to the decreased rotational correlation time of the Gd-dendrimer complex, in agreement with the literature.64–68 The relatively high r1 relaxivity is essential for the use of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes for sensitive MR imaging applications.Targeted MR imaging of cancer cells in vitro
With the proven targeting specificity of the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes to FAR-overexpressing cancer cells and the great r1 relaxivity of the complexes, we next explored the capability to use the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes for targeted MR imaging of cancer cells in vitro (Fig. 9). It can be seen that both KB-HFAR and KB-LFAR cells display a gradual MR contrast enhancement with the Gd concentration (Fig. 9a), suggesting that both cells are able to uptake the complexes. Quantitative analysis of the MR signal-to-noise ratio (SNR) reveals that MR SNR of both KB-HFAR and KB-LFAR cells treated with the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes are much higher than that of the corresponding cells treated with PBS, and higher Gd concentrations enable higher MR SNR of both cells (Fig. 9b). Importantly, under a given Gd concentration, the MR SNR of KB-HFAR cells is much higher than that of the KB-LFAR cells, especially at the high Gd concentrations (2 and 4 μM). This indicates that the FA-modification of dendrimers enables specific MR imaging of FAR-overexpressing cancer cells via an FA-mediated active targeting pathway. | ||
| Fig. 9 T1-weighted MR images (a) and MR SNRs (b) of KB-HFAR and KB-LFAR cells incubated with the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes at different Gd concentrations for 3 h (n = 3). | ||
In vitro cellular uptake assay
The targeted Gd uptake by KB-HFAR cells was further quantitatively confirmed by ICP-OES analysis (Fig. 10). It can be seen that at the studied Gd concentration (1 or 4 μM), the Gd uptake in KB-HFAR cells is much higher than that in KB-LFAR cells (p < 0.001). The ICP-OES data corroborate the in vitro MR imaging results, confirming the potential to use the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes as a nanoprobe for specific MR imaging of cancer cells.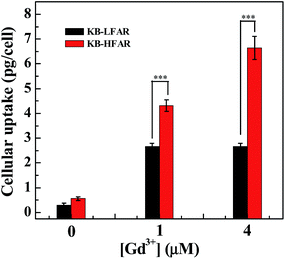 | ||
| Fig. 10 ICP-OES analysis of the Gd uptake in KB-HFAR and KB-LFAR cells after treated with the G5.NHAc–DOTA(Gd)–PEG–FA/DOX complexes at different Gd concentrations for 3 h. | ||
Conclusion
In summary, we developed a multifunctional dendrimeric theranostic nanoplatform for targeted imaging and therapy of cancer cells in vitro. The versatile dendrimer nanotechnology enables the modification of chelator/Gd complexes on the dendrimer periphery and physical encapsulation of DOX within the dendrimer interior. With the additional modification of targeting ligand FA via a PEG spacer, the developed dendrimeric theranostic nanoplatform with good colloidal stability can be used for targeted MR imaging and chemotherapy of FAR-overexpressing cancer cells due to the coexistence of the Gd(III)/chelator complex and the loaded drug DOX. With the ability to be modified with other targeting ligands, the developed dendrimeric nanoplatform may hold great promise to be used for theranostics of different types of cancer cells.Acknowledgements
This research is financially supported by the Program for New Century Excellent Talents in University, State Education Ministry, the National Natural Science Foundation of China (21273032), the Ph.D. Programs Foundation of Ministry of Education of China (20130075110004), the Sino-German Center for Research Promotion (GZ899), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, and the Chinese Universities Scientific Fund (CUSF-DH-D-2014017).Notes and references
- D. W. Bartlett, H. Su, I. J. Hildebrandt, W. A. Weber and M. E. Davis, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 15549–15554 CrossRef CAS PubMed.
- X. Huang, I. H. El-Sayed, W. Qian and M. A. El-Sayed, J. Am. Chem. Soc., 2006, 128, 2115–2120 CrossRef CAS PubMed.
- D. Kim, Y. Y. Jeong and S. Jon, ACS Nano, 2010, 4, 3689–3696 CrossRef CAS PubMed.
- J.-W. Kim, E. I. Galanzha, E. V. Shashkov, H.-M. Moon and V. P. Zharov, Nat. Nanotechnol., 2009, 4, 688–694 CrossRef CAS PubMed.
- J. R. McCarthy, F. A. Jaffer and R. Weissleder, Small, 2006, 2, 983–987 CrossRef CAS PubMed.
- N. Nasongkla, E. Bey, J. Ren, H. Ai, C. Khemtong, J. S. Guthi, S.-F. Chin, A. D. Sherry, D. A. Boothman and J. Gao, Nano Lett., 2006, 6, 2427–2430 CrossRef CAS PubMed.
- N. Rapoport, Z. Gao and A. Kennedy, J. Natl. Cancer Inst., 2007, 99, 1095–1106 CrossRef CAS PubMed.
- S. Santra, C. Kaittanis, J. Grimm and J. M. Perez, Small, 2009, 5, 1862–1868 CrossRef CAS PubMed.
- M. E. Davis and D. M. Shin, Nat. Rev. Drug Discovery, 2008, 7, 771–782 CrossRef CAS PubMed.
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nat. Nanotechnol., 2007, 2, 751–760 CrossRef CAS PubMed.
- H. Li, Y. Jia, A. Wang, W. Cui, H. Ma, X. Feng and J. Li, Chem.–Eur. J., 2014, 20, 499–504 CrossRef CAS PubMed.
- Y. Yang, Y. Jia, L. Gao, J. Fei, L. Dai, J. Zhao and J. Li, Chem. Commun., 2011, 47, 12167–12169 RSC.
- Y. Yang and J. Li, Adv. Colloid Interface Sci., 2014, 207, 155–163 CrossRef CAS PubMed.
- Y. Yang, W. Song, A. Wang, P. Zhu, J. Fei and J. Li, Phys. Chem. Chem. Phys., 2010, 12, 4418–4422 RSC.
- Y. Yang, X. Yan, Y. Cui, Q. He, D. Li, A. Wang, J. Fei and J. Li, J. Mater. Chem., 2008, 18, 5731–5737 RSC.
- L. Brannon-Peppas and J. O. Blanchette, Adv. Drug Delivery Rev., 2012, 64, 206–212 CrossRef PubMed.
- K. Park, S. Lee, E. Kang, K. Kim, K. Choi and I. C. Kwon, Adv. Funct. Mater., 2009, 19, 1553–1566 CrossRef CAS.
- T. Grobner and F. Prischl, Kidney Int., 2007, 72, 260–264 CrossRef CAS PubMed.
- J. Wang, D. Short, N. Sebire, I. Lindsay, E. Newlands, P. Schmid, P. Savage and M. Seckl, Ann. Oncol., 2008, 19, 1578–1583 CrossRef CAS PubMed.
- J. R. McCarthy, Nanomedicine, 2009, 4, 693–695 CrossRef PubMed.
- B. Sumer and J. M. Gao, Nanomedicine, 2008, 3, 137–140 CrossRef PubMed.
- J. Li, Y. Hu, J. Yang, P. Wei, W. Sun, M. Shen, G. Zhang and X. Shi, Biomaterials, 2015, 38, 10–21 CrossRef CAS PubMed.
- Z. Zhou, B. Kong, C. Yu, X. Shi, M. Wang, W. Liu, Y. Sun, Y. Zhang, H. Yang and S. Yang, Sci. Rep., 2014, 4, 3653 Search PubMed.
- M. Shen and X. Shi, Nanoscale, 2010, 2, 1596–1610 RSC.
- J. Zhu and X. Shi, J. Mater. Chem. B, 2013, 1, 4199–4211 RSC.
- Z. Qiao and X. Shi, Prog. Polym. Sci., 2014 DOI:10.1016/j.progpolymsci.2014.1008.1002.
- D. A. Tomalia and J. M. J. Frechet, Dendrimers and Other Dendritic Polymers, John Wiley & Sons Ltd, New York, 2001 Search PubMed.
- J. F. Kukowska-Latallo, K. A. Candido, Z. Cao, S. S. Nigavekar, I. J. Majoros, T. P. Thomas, L. P. Balogh, M. K. Khan and J. R. Baker Jr, Cancer Res., 2005, 65, 5317–5324 CrossRef CAS PubMed.
- I. J. Majoros, A. Myc, T. Thomas, C. B. Mehta and J. R. Baker Jr, Biomacromolecules, 2006, 7, 572–579 CrossRef CAS PubMed.
- I. J. Majoros, T. P. Thomas, C. B. Mehta and J. R. Baker Jr, J. Med. Chem., 2005, 48, 5892–5899 CrossRef CAS PubMed.
- T. P. Thomas, I. J. Majoros, A. Kotlyar, J. F. Kukowska-Latallo, A. Bielinska, A. Myc and J. R. Baker Jr, J. Med. Chem., 2005, 48, 3729–3735 CrossRef CAS PubMed.
- Y. Zheng, F. Fu, M. Zhang, M. Shen, M. Zhu and X. Shi, MedChemComm, 2014, 5, 879–885 RSC.
- X. Shi, I. Lee, X. Chen, M. Shen, S. Xiao, M. Zhu, J. R. Baker Jr and S. H. Wang, Soft Matter, 2010, 6, 2539–2545 RSC.
- Y. Wang, X. Cao, R. Guo, M. Shen, M. Zhang, M. Zhu and X. Shi, Polym. Chem., 2011, 2, 1754–1760 RSC.
- Y. Wang, R. Guo, X. Cao, M. Shen and X. Shi, Biomaterials, 2011, 32, 3322–3329 CrossRef CAS PubMed.
- M. Zhang, R. Guo, Y. Wang, X. Cao, M. Shen and X. Shi, Int. J. Nanomed., 2011, 6, 2337–2349 CAS.
- M. Zhang, R. Guo, M. Kéri, I. Bányai, Y. Zheng, M. Cao, X. Cao and X. Shi, J. Phys. Chem. B, 2014, 118, 1696–1706 CrossRef CAS PubMed.
- H. Wang, L. Zheng, C. Peng, R. Guo, M. Shen, X. Shi and G. Zhang, Biomaterials, 2011, 32, 2979–2988 CrossRef CAS PubMed.
- C. Peng, L. Zheng, Q. Chen, M. Shen, R. Guo, H. Wang, X. Cao, G. Zhang and X. Shi, Biomaterials, 2012, 33, 1107–1119 CrossRef CAS PubMed.
- H. Liu, Y. Xu, S. Wen, Q. Chen, L. Zheng, M. Shen, J. Zhao, G. Zhang and X. Shi, Chem.–Eur. J., 2013, 19, 6409–6416 CrossRef CAS PubMed.
- C. Peng, K. Li, X. Cao, T. Xiao, W. Hou, L. Zheng, R. Guo, M. Shen, G. Zhang and X. Shi, Nanoscale, 2012, 4, 6768–6778 RSC.
- Q. Chen, K. Li, S. Wen, H. Liu, C. Peng, H. Cai, M. Shen, G. Zhang and X. Shi, Biomaterials, 2013, 34, 5200–5209 CrossRef CAS PubMed.
- S. Wen, K. Li, H. Cai, Q. Chen, M. Shen, Y. Huang, C. Peng, W. Hou, M. Zhu and G. Zhang, Biomaterials, 2013, 34, 1570–1580 CrossRef CAS PubMed.
- Y. Cao, Y. He, H. Liu, Y. Luo, M. Shen, J. Xia and X. Shi, J. Mater. Chem. B, 2015, 3, 286–295 RSC.
- H. Liu, H. Wang, Y. Xu, M. Shen, J. Zhao, G. Zhang and X. Shi, Nanoscale, 2014, 6, 4521–4526 RSC.
- L. Zheng, J. Zhu, M. Shen, X. Chen, J. R. Baker Jr, S. H. Wang, G. Zhang and X. Shi, MedChemComm, 2013, 4, 1001–1005 RSC.
- Y. Chang, X. Meng, Y. Zhao, K. Li, B. Zhao, M. Zhu, Y. Li, X. Chen and J. Wang, J. Colloid Interface Sci., 2011, 363, 403–409 CrossRef CAS PubMed.
- J. Y. Zhu, L. F. Zheng, S. H. Wen, Y. Q. Tang, M. W. Shen, G. X. Zhang and X. Y. Shi, Biomaterials, 2014, 35, 7635–7646 CrossRef CAS PubMed.
- J. Li, Y. He, W. Sun, Y. Luo, H. Cai, Y. Pan, M. Shen, J. Xia and X. Shi, Biomaterials, 2014, 35, 3666–3677 CrossRef CAS PubMed.
- Y. Yang, Y. Sun, T. Cao, J. Peng, Y. Liu, Y. Wu, W. Feng, Y. Zhang and F. Li, Biomaterials, 2013, 34, 774–783 CrossRef CAS PubMed.
- X. He, C. Alves, N. Oliveira, J. Rodrigues, J. Zhu, I. Bányai, H. Tomás and X. Shi, Colloids Surf., B, 2015, 125, 82–89 CrossRef CAS PubMed.
- H. Liao, H. Liu, Y. Li, M. Zhang, H. Tomás, M. Shen and X. Shi, J. Appl. Polym. Sci., 2014, 131, 40358 Search PubMed.
- C. Kojima, K. Kono, K. Maruyama and T. Takagishi, Bioconjugate Chem., 2000, 11, 910–917 CrossRef CAS PubMed.
- F. Fu, Y. Wu, J. Zhu, S. Wen, M. Shen and X. Shi, ACS Appl. Mater. Interfaces, 2014, 6, 16416–16425 CAS.
- X. Shi, I. Banyai, K. Rodriguez, M. T. Islam, W. Lesniak, P. Balogh, L. P. Balogh and J. R. Baker Jr, Electrophoresis, 2006, 27, 1758–1767 CrossRef CAS PubMed.
- D. Cakara, J. Kleimann and M. Borkovec, Macromolecules, 2003, 36, 4201–4207 CrossRef CAS.
- K. Kono, C. Kojima, N. Hayashi, E. Nishisaka, K. Kiura, S. Wataral and A. Harada, Biomaterials, 2008, 29, 1664–1675 CrossRef CAS PubMed.
- I. G. Campbell, T. A. Jones, W. D. Foulkes and J. Trowsdale, Cancer Res., 1991, 51, 5329–5338 CAS.
- J. F. Ross, P. K. Chaudhuri and M. Ratnam, Cancer, 1994, 73, 2432–2443 CrossRef CAS.
- Y.-J. Lu, K.-C. Wei, C.-C. M. Ma, S.-Y. Yang and J.-P. Chen, Colloids Surf., B, 2012, 89, 1–9 CrossRef CAS PubMed.
- H. Yao, S. S. Ng, W. O. Tucker, Y.-K.-T. Tsang, K. Man, X.-m. Wang, B. K. C. Chow, H.-F. Kung, G.-P. Tang and M. C. Lin, Biomaterials, 2009, 30, 5793–5803 CrossRef CAS PubMed.
- S. Wang, Y. Wu, R. Guo, Y. Huang, S. Wen, M. Shen, J. Wang and X. Shi, Langmuir, 2013, 29, 5030–5036 CrossRef CAS PubMed.
- Y. Wu, R. Guo, S. Wen, M. Shen, M. Zhu, J. Wang and X. Shi, J. Mater. Chem. B, 2014, 2, 7410–7418 RSC.
- S. D. Swanson, J. F. Kukowska-Latallo, A. K. Patri, C. Chen, S. Ge, Z. Cao, A. Kotlyar, A. T. East and J. R. Baker Jr, Int. J. Nanomed., 2008, 3, 201–210 CrossRef CAS PubMed.
- L. H. Bryant, M. W. Brechbiel, C. C. Wu, J. W. M. Bulte, V. Herynek and J. A. Frank, J. Magn. Reson. Imaging, 1999, 9, 348–352 CrossRef.
- S. Laus, A. Sour, R. Ruloff, E. Toth and A. E. Merbach, Chem.–Eur. J., 2005, 11, 3064–3076 CrossRef CAS PubMed.
- G. M. Nicolle, E. Toth, H. Schmitt-Willich, B. Raduchel and A. E. Merbach, Chem.–Eur. J., 2002, 8, 1040–1048 CrossRef CAS.
- J. Rudovsky, M. Botta, P. Hermann, K. I. Hardcastle, I. Lukes and S. Aime, Bioconjugate Chem., 2006, 17, 975–987 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional experimental results. See DOI: 10.1039/c5ra01215e |
| This journal is © The Royal Society of Chemistry 2015 |