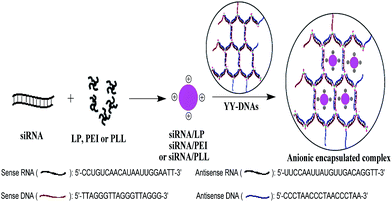
Delivery of siRNA using siRNA/cationic vector complexes encapsulated in dendrimer-like polymeric DNAs
Ahmed F. M. El-Mahdy*ab,
Takayuki Shibataa,
Tsutomu Kabashimaa,
Qinchang Zhua and
Masaaki Kai*a
aFaculty of Pharmaceutical Sciences, Graduate School of Biomedical Sciences, Nagasaki University, Nagasaki 852-8521, Japan. E-mail: ahmed.ahmed20@science.au.edu.eg; ms-kai@nagasaki-u.ac.jp; Fax: +81-95-819-2438
bChemistry Department, Faculty of Science, Assiut University, Assiut 71516, Egypt
First published on 27th March 2015
Abstract
Small interfering RNA (siRNA) is a powerful gene silencing tool and has been considered a potential agent for the treatment of many diseases. However, development of safe and effective siRNA delivery systems still remains a great challenge. In this study, we developed a new siRNA delivery system based on the electrostatic encapsulation of siRNA/cationic vector complexes with dendrimer-like polymeric DNAs (YY-DNAs). The binary complexes of siRNA with cationic vectors such as lipofectamine (LP), polyethylenimine (PEI) and poly-L-lysine (PLL) were first constructed. Then, the encapsulation was performed by the addition of YY-DNAs to the binary complexes in order to form stable complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs. The encapsulated siRNA complexes showed nearly spherical morphology with about 13–37 nm average hydrodynamic size and their ζ-potentials were negative. The cationic complexes of siRNA/LP, siRNA/PEI and siRNA/PLL showed a high cytotoxicity towards the cells and strong aggregation with erythrocytes, while their encapsulation into YY-DNAs dramatically decreased the toxicities of complexes. Furthermore, these anionic encapsulated siRNA complexes were highly taken up by the HeLa cells and showed extremely high cellular uptake efficiencies and gene silencing effects without such cytotoxicity and aggregation. The stability of these complexes in 10% FBS and human serum was investigated and they showed high stability even after incubation for 72 h and 48 h, respectively. Therefore, we have newly identified safe and efficient anionic complexes of siRNA for clinical uses.
1. Introduction
Small interfering RNA (siRNA) is a class of double-stranded RNA molecules, 20–25 base pair in length, which is able to induce RNA interference (RNAi). Since its first report in 1998, RNAi has been considered a potential agent for the treatment of many diseases, including cancer, viral infections and hereditary genetic disorders.1,2 In the general RNAi process, long transcripts of double-stranded RNA (dsRNA) are cleaved into small interfering RNAs (siRNAs) by the help of an endoribonuclease dicer. The resulting siRNA molecule is then loaded onto the RNA-induced silencing complex (RISC) to form RISC–siRNA complex. After activation, the siRNA is unwrapped and one of the two strands is released, resulting in an activated form of RISC with a single-stranded RNA. This RISC–RNA complex then binds to mRNA homologous in sequence to the siRNA by base-pairing recognition and guides its sequence-specific degradation and consequently knocks down the expression of the corresponding protein. This procedure is also known as gene silencing that is highly effective and specific, because one nucleotide mismatch between the target mRNA and the siRNA can prevent the recognition and thus the silencing process.3The clinical application of siRNA in disease therapy faces significant challenges such as degradation by endogenous enzymes and low cellular uptake. Therefore, successful development of RNAi for clinical applications depends on adequate delivery systems that can efficiently protect and accumulate siRNA molecules in target cells and tissues.4 In siRNA delivery, virus-derived carriers have shown high efficiency to deliver siRNA to host cells by taking advantage of intracellular trafficking machineries. However, due to several drawbacks such as high cost of production and safety concerns, non-viral siRNA vectors have attracted more and more attentions. These vectors typically possess cationic nature (e.g., cationic cell penetrating peptides, cationic polymers, dendrimers, and cationic lipids) and complex with siRNA by electrostatic interaction.5 For efficient in vivo siRNA delivery to target cells by these cationic vectors, the cationic vector/siRNA complex must be stabilized in the blood by avoiding its agglutination with blood components. This is because the electrostatic interactions between positively charged vector and negatively charged erythrocytes cause agglutination,6 and thus contribute to high entrapment of vectors in the highly extended body capillaries.7
Recently, one promising approach for overcome this problem which applied for gene delivery, is the electrostatic encapsulation of cationic vector with anionic polymers such as γ-poly-L-glutamic acid (γ-PGA),8 hyaluronic acid (HA)9 and chondroitin sulphate (CS).10 These anionic polymers display a great number of attractive properties, including preventing of agglutination with the blood components, preparation of very small vesicles, better transfection efficacy and better target specificity.11,12
Anionic polymers have been already been developed for plasmid DNA (pDNA) delivery, while there is little information about their using for siRNA delivery. In addition, the in vitro transfection of siRNA need prolonged incubation time than that of pDNA and thus the use of safe and efficient delivery system of siRNA is often needed. Furthermore, the in vivo siRNA delivery system for targeting organs and cells has received significant attention in clinical applications. Therefore, in this study, we constructed a safe and efficient siRNA delivery system depended on the electrostatic encapsulation of siRNA/cationic vector complexes with dendrimer-like polymeric DNAs (YY-DNAs), in order to form stable anionic particles of siRNA/lipofectamine (LP)/YY-DNAs, siRNA/polyethylenimine (PEI)/YY-DNAs and siRNA/poly-L-lysine (PLL)/YY-DNAs, respectively (Scheme 1).
The advantages of the using of our synthesized dendrimer-like polymeric DNAs (YY-DNAs) to coated the cationic binary complexes is that our YY-DNAs is synthesized mainly from DNA strands that are one of the body components, in order to form highly safe siRNA delivery systems. While, all reported anionic polymers such as γ-PGA, HA and CS are chemically composed polymers,8–10,13–15 which have toxicity on cells. In addition, the YY-DNAs is to easily prepare it by using the maleimide–thiol coupling for efficient synthesis of Y-shaped DNAs by just incubation of sense and antisense DNAs with a thiol group at 5′-end and tris(2-maleimidoethyl)amine (TMEA) at room temperature for overnight followed by self-assembly of them at 37 °C for 1 h (Scheme 2). On the other hand, the reported anionic polymers were synthesized via many reaction steps and needed longer time. Therefore, our system for siRNA delivery is more safety, time-saving and different from the previously reported ones.
2. Experimental
2.1. Materials
Polyethylenimine (PEI, branched form with average molecular weight of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) was supplied from Aldrich Chemical Co. (Milwaukee, USA). Lipofectamine® 2000 (LP) was purchased from Invitrogen Life Technologies (New York, USA). Poly-L-lysine (PLL) was obtained from Sigma (St. Louis, MO, USA), Dulbecco's Modified Eagle Medium (D-MEM) was obtained from Wako Pure Chemical Industries (Osaka, Japan). Fetal bovine serum (FBS) was obtained from thermo scientific (Tokyo, Japan). Antibiotics (10
000) was supplied from Aldrich Chemical Co. (Milwaukee, USA). Lipofectamine® 2000 (LP) was purchased from Invitrogen Life Technologies (New York, USA). Poly-L-lysine (PLL) was obtained from Sigma (St. Louis, MO, USA), Dulbecco's Modified Eagle Medium (D-MEM) was obtained from Wako Pure Chemical Industries (Osaka, Japan). Fetal bovine serum (FBS) was obtained from thermo scientific (Tokyo, Japan). Antibiotics (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units penicillin, 10 mg streptomycin and 25 μg amphotericin B per mL) was supplied from Nacalai Tesque Inc. (Tokyo, Japan). Trypsin was acquired from GIBCO BRL (Grand Island, NY, USA). 5′-Thiol-blocked sense single strand (ss) DNA (HO–(CH2)6–S–S–(CH2)6–5′-TTAGGGTTAGGGTTAGGG-3′), 5′-thiol-blocked-3′-FAM-labeled sense ssDNA (HO–(CH2)6–S–S–(CH2)6–5′-TTAGGGTT-AGGGTTAGGG-3′-FAM) and 5′-thiol-blocked antisense ssDNA (HO–(CH2)6–S–S–(CH2)6–5′-CCCTAACCCTAACCC-TAA-3′) were acquired from Takara Shuzo Co., Ltd. (Otsu, Shiga, Japan). HIV-1 PR siRNA (sense: 5′-CCUGUCAACAU-AAUUGGAATT-3′; antisense: 5′-UUCCAAUUAUGUUGAC-AGGTT-3′) was acquired from Sigma Genosys Japan (Ishikari, Japan). Cell Counting Kit-8 (CCK-8) was obtained from Dojindo Molecular Technologies, Inc. (Kumamoto, Japan). All chemicals were obtained of analytical grade and were used without further purification.
000 units penicillin, 10 mg streptomycin and 25 μg amphotericin B per mL) was supplied from Nacalai Tesque Inc. (Tokyo, Japan). Trypsin was acquired from GIBCO BRL (Grand Island, NY, USA). 5′-Thiol-blocked sense single strand (ss) DNA (HO–(CH2)6–S–S–(CH2)6–5′-TTAGGGTTAGGGTTAGGG-3′), 5′-thiol-blocked-3′-FAM-labeled sense ssDNA (HO–(CH2)6–S–S–(CH2)6–5′-TTAGGGTT-AGGGTTAGGG-3′-FAM) and 5′-thiol-blocked antisense ssDNA (HO–(CH2)6–S–S–(CH2)6–5′-CCCTAACCCTAACCC-TAA-3′) were acquired from Takara Shuzo Co., Ltd. (Otsu, Shiga, Japan). HIV-1 PR siRNA (sense: 5′-CCUGUCAACAU-AAUUGGAATT-3′; antisense: 5′-UUCCAAUUAUGUUGAC-AGGTT-3′) was acquired from Sigma Genosys Japan (Ishikari, Japan). Cell Counting Kit-8 (CCK-8) was obtained from Dojindo Molecular Technologies, Inc. (Kumamoto, Japan). All chemicals were obtained of analytical grade and were used without further purification.
2.2. Synthesis of FAM-labeled dendrimer-like polymeric DNAs (FAM-YY-DNAs)
Dendrimer-like polymeric DNAs (YY-DNAs) was synthesized according to our recently method.16,17 By the same manner, we synthesized a new FAM-labeled dendrimer-like polymeric DNAs (FAM-YY-DNAs). 3′-FAM-labeled sense and antisense ssDNAs possessing a free thiol group at 5′-end were prepared as follows: 125 μL of 100 mM DTT in sodium phosphate buffer (pH 8.5) was mixed with 15.24 nmol of thiol-blocked sense (100 μg) or antisense (84.24 μg) ssDNA at the 5′-end, and incubated at room temperature for 1 h to produce free thiol group. After incubation, the excess DTT and other byproducts were removed by using a NAP-10 column and the concentration was determined by measuring the absorbance at 260 nm.For the preparation of FAM-labeled Y-DNA and Y-cDNA: 15.6 nmol of sense free 5′-thiol-3′-FAM (100 μg) or its antisense free 5′-thiol ssDNA (85.7 μg) in 100 μL of 1× PBS (pH 7.4) was reacted with 5.2 nmol of tris(2-maleimidoethyl)amine (TMEA)16,17 (2 μg) dissolved in 6 μL of acetone. After overnight incubation at room temperature, the residual TMEA and acetone were eliminated through the NAP-10 column. The products of FAM-labeled Y-DNA and Y-cDNA were analyzed by 20% polyacrylamide gel electrophoresis (PAGE) (Fig. 1). The gel was stained with a fluorescent dye, SYBR-Gold (Invitrogen™, Eugene, USA). Each band intensity was calculated by a densitometry using Image J software (National Institute of Health, USA).
Equal amounts of each resulting product of FAM-labeled Y-DNA (39.8 μg, 3 nmol) and Y-cDNA (35.78 μg, 3 nmol) were assembled by incubation in 1× PBS buffer (pH 7.4) at 37 °C for 1 h to yield 1.5 nmol of the FAM-labeled dendrimer-like polymeric DNAs (FAM-YY-DNAs) (75.85 μg). The product of FAM-YY-DNAs was analyzed by 20% polyacrylamide gel electrophoresis (PAGE) (Fig. 1). The gel was stained with a fluorescent dye, SYBR-Gold (Invitrogen™, Eugene, USA).
2.3. Electrostatic encapsulation of siRNA binary complexes with YY-DNAs
The cationic siRNA binary complexes (siRNA/LP, siRNA/PEI and siRNA/PLL) were prepared by mixing a solution of siRNA (1 μg) in 8 μL of 1× PBS (pH 7.4) with a solution of LP, PEI or PLL (8 μg) in 8 μL of 1× PBS (pH 7.4) at a weight ratio (w![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) w) of 1
w) of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 and then left for 30 min at room temperature.
8 and then left for 30 min at room temperature.
The anionic encapsulated siRNA complexes (siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs) were prepared by adding different amounts of the anionic YY-DNAs (2–8 μg) in 14 μL of 1× PBS (pH 7.4) to each of the formed cationic siRNA binary complex and left for additional 30 min at room temperature.
2.4. Morphology, particle size and zeta potential
The morphology of the cationic siRNA binary complexes and various anionic encapsulated siRNA complexes were studied by scanning electron microscopy (SEM), using JSM-840 SEM (JEOL Ltd., Tokyo, Japan). The samples in water were placed on a silicon chip without any dopant and then allowed to dry at room temperature under desiccator. The silicon chip was then gold–palladium coated using an auto fine coater (JEOL JFC-1600, JEOL, Tokyo, Japan). They were then observed under SEM.The particle size and ζ-potential of the cationic siRNA binary complexes and the anionic encapsulated siRNA complexes were measured at 25 °C using a Zetasizer Nano ZS (Malven Instruments, Worcs, UK) at 90° to the incident beam (623 nm wavelength from 4 mW He–Ne laser tubes) in 1 cm length quartz cuvette. Data fitting was carried out using a multimodal algorithm supplied by Malvern Instruments and then the hydrodynamic diameter was identified using the Einstein–Stockes equation. Each measurement was performed in triplicate.
2.5. Agarose gel retardation assay
The gel retardation assay of various anionic encapsulated siRNA complexes was evaluated by using agarose gel electrophoresis. A portion of 15 μL of each complex was mixed with 1 μL of 6× loading buffer and then loaded onto 3% agarose gel. Electrophoresis was carried out at 100 V for 30 min in Tris–borate–EDTA (pH 8.0) buffer and the complexes were visualized by ethidium bromide staining.2.6. In vitro release of siRNA from the anionic encapsulated complexes
To assess siRNA release profile, anionic encapsulate complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and containing 5 μg of FAM-labeled siRNA, were prepared as described above. Each anionic complex was incubated at 37 °C in 1 mL of 1× PBS (pH 7.4) with moderate shaking. At time points ranging from 1 to 7 days, the supernatant were collected by centrifugation at 14
4 and containing 5 μg of FAM-labeled siRNA, were prepared as described above. Each anionic complex was incubated at 37 °C in 1 mL of 1× PBS (pH 7.4) with moderate shaking. At time points ranging from 1 to 7 days, the supernatant were collected by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. Then, the amount of released siRNA was determined by measuring the fluorescence intensity of the supernatant at an emission wavelength of 520 nm with an excitation of 495 nm, using a FP-8200 spectrofluorometer (JASCO, Tokyo, Japan). The released siRNA (%) was calculated as a ratio (%) of the released amount, to the total amount of siRNA.
000 rpm for 10 min. Then, the amount of released siRNA was determined by measuring the fluorescence intensity of the supernatant at an emission wavelength of 520 nm with an excitation of 495 nm, using a FP-8200 spectrofluorometer (JASCO, Tokyo, Japan). The released siRNA (%) was calculated as a ratio (%) of the released amount, to the total amount of siRNA.
2.7. Cell culture
HeLa cells were purchased from Riken BRC Cell Bank (Tsukuba, Japan). Cells were cultured in Dulbecco's Modified Eagle Medium (D-MEM) medium supplemented with 10% fetal bovine serum (FBS), 100 U mL−1 penicillin, 100 μg mL−1 streptomycin and 0.25 μg mL−1 amphotericin B at 37 °C under a humidified atmosphere containing 5% CO2.2.8. Cytotoxicity assay
Cytotoxicity of various cationic siRNA binary complexes and anionic encapsulated siRNA complexes were measured using cell counting kit-8 (CCK-8) assay. HeLa cells were seeded into a 96-well plate at a density of 5 × 103 cells per well, and cultured for 24 h in 100 μL of D-MEM medium supplementary with 10% FBS at 37 °C in a humidified atmosphere of 5% CO2. A portion of 10 μL of cationic siRNA binary complex or anionic encapsulated siRNA complex containing 2 μg of siRNA at various weight ratios was added and incubated for further 24 h. After 10 μL of CCK-8 solution was added to each well, the cells were incubated for an additional 3 h. The absorbance at 450 nm was recorded by using UV-Vis spectrometer (Jasco V-630 Bio spectrophotometer, Tokyo, Japan). All experiments were conducted in triplicate.2.9. Interaction with erythrocytes
Erythrocytes from mice were washed three times with 1 mL of 1× PBS (pH 7.4) at 4 °C by centrifugation at 3000 g (eppendorf centrifuge 5415R, Hamburg, Germany) for 10 min. A 2% (v/v) stock suspension of erythrocytes in PBS was prepared for aggregation study. A portion of 38 μL of various anionic encapsulated siRNA complexes in 1× PBS containing 2 μg of siRNA were added to 78 μL of the stock suspension of erythrocytes and incubated for 15 min at 37 °C. Then 10 μL of the suspension was placed on a glass plate and the aggregation was observed by microscopy (400× magnification). For the turbidity study, the turbidity was quantified using UV-Vis spectrometer (Jasco V-630 Bio spectrophotometer, Tokyo, Japan) by measuring the absorbance at a wavelength of 630 nm. The results are shown as a percentage of siRNA/LP.2.10. Cellular uptake of anionic encapsulated siRNA complexes
To visualize the uptake of various cationic siRNA binary complexes and anionic encapsulated siRNA complexes, HeLa cells at a density of 2 × 105 cells per well were cultured in 100 μL of D-MEM medium supplementary with 10% FBS at 37 °C. After 24 h incubation, the medium was replaced with 100 μL of fresh D-MEM medium and each siRNA binary complex or encapsulated siRNA complex containing 2 μg of FAM-labeled siRNA was separately added to the cells and incubated for 4 h. Then the cells were washed three times with D-MEM medium and observed under bright-field optical microscopy (EVOS, AME-3302, Bothell, WA) and inverted fluorescence microscopy (LEICA DMIRB, Wetzlar, Germany).To determine the uptake of various cationic siRNA binary complexes and anionic encapsulated siRNA complexes, HeLa cells were transferred into a 0.5 mL eppendorf tube at a density of 2 × 105 cells per tube, and cultured in 100 μL of D-MEM medium supplementary with 10% FBS at 37 °C with shaking perpendicularly. After 24 h incubation, the medium was replaced with 100 μL of fresh D-MEM medium by centrifugation at 1000 g for 5 min and each siRNA binary complex or encapsulated siRNA complex containing 2 μg of FAM-labeled siRNA was separately added to the cells and incubated for 4 h with shaking perpendicularly. The cells were washed two times with 200 μL of PBS buffer and lysed in 200 μL of sterile H2O by vortex and sonicated three times for 10 min in ice. Then the lysate supernatant was collected by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200g for 10 min. The total protein concentration in the lysate was determined by the Kit of Quick StartTM Bradford Protein Assay (Bio-Red, CA, USA). The fluorescence intensity of each lysate solution was measured at an emission wavelength of 520 nm with an excitation of 495 nm, using a FP-8200 spectrofluorometer (JASCO, Tokyo, Japan).
200g for 10 min. The total protein concentration in the lysate was determined by the Kit of Quick StartTM Bradford Protein Assay (Bio-Red, CA, USA). The fluorescence intensity of each lysate solution was measured at an emission wavelength of 520 nm with an excitation of 495 nm, using a FP-8200 spectrofluorometer (JASCO, Tokyo, Japan).
2.11. Gene expression analysis
HeLa cells were seeded at density of 2 × 105 cells per well in a 12-well plate and cultivated in 1 mL D-MEM medium supplementary with 10% FBS under 37 °C and 5% CO2 overnight. Prior to transfection, medium was exchanged with 1 mL of fresh medium when cells reach an 80–90% confluence. Then 200 μL of D-MEM medium containing 1.5 μg of pcDNA-HIV-PR and 3 μL of LP, PEI or PLL was added in each well. After transfection for 4 h, the cells were washed three times with 500 μL of D-MEM medium to remove the residue transfection reagent. Thereafter, various cationic siRNA binary or anionic encapsulated siRNA complexes containing 1.5 μg of HIV-1 PR siRNA into 1.2 mL of D-MEM medium were added to the cells. Finally, the cells were harvested 48 h after the transfection of HIV-PR-expressing plasmid and analyzed for the inhibition of HIV-PR mRNA expression by real-time RT-PCR (Smat Cycler II System, Cepheid, USA) with a SYBR Premix Ex Taq_kit.Briefly, total cellular RNA was extracted with Sepasol RNA I Super G (Nacalai Tesque, Japan) and treated with 2 unit of DNase I to eliminate potential DNA contamination. The RNA was then reverse transcribed into cDNA using PrimeScript RT reagent Kit (TaKaRa, Japan). The quantity of HIV-PR cDNA was finally determined by the real-time PCR with an HIV-PR-specific primer pair, HIV-PR-F (5′-TCACTCTTTGGCAACG-ACCC-3′) and HIV-PR-R (5′-TTCCATCTTCCTGGCAAACA-AACTC-3′). The endogenous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the normalizing gene (GAPDH-F 5′-AAGGTCGGAGTCAACGGATT-3′ and GAPDH-R 5′-CTGGAAGATGGTGATGGGATT-3′). The expression yield of HIV-PR mRNA in treated and control cells were determined according to the normalized threshold cycle values, using the formula 2−ΔΔCt, where ΔΔCt = (Ct,HIV-PR − Ct,GAPDH)treat − (Ct,HIV-PR − Ct,GAPDH)control.18
2.12. Stability study of encapsulated siRNA complex
Anionic encapsulated siRNA complex containing 2 μg of siRNA was incubated at 37 °C in 200 μL of D-MEM medium containing 10% FBS, or in human serum. Aliquots of 2 pmol of them were withdraw at predetermined time points and immediately frozen in 1% diethylpyrocarbonate (DEPC). The time point of 0 h was that sample was collected immediately after mixing the complex with 10% FBS medium or human serum. All samples were subjected to electrophoresis with 3% agarose gel and visualized by staining with ethidium bromide.3. Results and discussion
3.1. Synthesis of FAM-labeled dendrimer-like polymeric DNAs (FAM-YY-DNAs)
In our recently studies,16,17 we reported the synthesis of dendrimer-like polymeric DNAs (YY-DNAs) by hybridization between sense Y-shaped single strand (ss) DNA (Y-DNA) and its antisense Y-shaped ssDNA (Y-cDNA) containing Y-shaped branches as a cross-linkers. Continuously, in the present study, we synthesized FAM-labeled dendrimer-like polymeric DNAs (FAM-YY-DNAs) by the same method as shown in Scheme 2. 3′-FAM-labeled sense and antisense ssDNAs possessing a thiol group at 5′-end were reacted with a synthesized trimeric cross-linker, tris(2-maleimidoethyl)amine (TMEA)16,17 after reduction and deprotection of the thiol group at 5′-end with dithiotheritol (DTT), in order to yield FAM-labeled sense Y-shaped ssDNA (FAM-Y-DNA) and its antisense Y-shaped ssDNA (Y-cDNA), respectively. The resulting products contained sense and antisense Y-shaped trimeric, dimeric and monomeric DNAs which were separated on 20% PAGE (Fig. 1). The sense FAM-Y-DNA is composed of 15% trimers, 60% dimers and 25% monomers, which were determined by the calculation of each band intensity using Image J software. The antisense Y-cDNA which was synthesized in the same manner, is composed of 28% trimers, 50% dimers and 22% monomers.Equal amounts of the sense FAM-Y-DNA and antisense Y-cDNA in a phosphate buffered saline (1× PBS) solution (pH 7.4) could hybridize with each other to produce the dendrimer FAM-YY-DNAs (Scheme 2) which was analyzed using 20% PAGE, as shown in Fig. 1.
3.2. Physicochemical characteristics and agarose gel retardation assay
Cationic vectors including liposomes,19 cyclodextrin,20 PEI21 and peptides22 had been used as cationic carriers for delivery system of siRNA in animal studies by several research groups. Here, we selected a commercially available transfection reagent, Lipofectamine® 2000 (LP) and two previously reported transfection reagents polyethylenimine (PEI) and poly-L-lysine (PLL), for the formation of cationic siRNA complexes of siRNA/LP, siRNA/PEI and siRNA/LP, respectively. Then the formed siRNA binary complexes were electrostatically encapsulated with our synthesized dendrimer-like polymeric DNAs (YY-DNAs), to form anionic encapsulated complexes that might be used for safe and efficacy siRNA delivery (Scheme 1).First, it is well known that cationic vectors complex siRNA via electrostatic interaction. The formation of cationic siRNA/LP, siRNA/PEI and siRNA/PLL binary complexes at different weight ratio was monitored by gel retardation assay. As shown in Fig. 2A, C and E, siRNA was detected as a band on agarose gel. The slight bands of siRNA were detected at weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and then no bands were observed at weight ratio of 1
2 and then no bands were observed at weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and 1
4 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8, indicated that siRNA completely bound to cationic carriers and formed stable binary complexes at weight ratio of 1
8, indicated that siRNA completely bound to cationic carriers and formed stable binary complexes at weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 or 1
4 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8. According to our results, manufacturer's protocol of LP, and previously research on PEI23 which mentioned that the siRNA/PEI with a ratio of 1
8. According to our results, manufacturer's protocol of LP, and previously research on PEI23 which mentioned that the siRNA/PEI with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 had highest transgene efficiency under in vitro and in vivo conditions, we chosen the cationic siRNA binary complexes with a weight ratio of 1
8 had highest transgene efficiency under in vitro and in vivo conditions, we chosen the cationic siRNA binary complexes with a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 for the further studies.
8 for the further studies.
 | ||
| Fig. 2 Gel retardation assays of (A) siRNA/LP, (B) siRNA/LP/YYDNAs, (C) siRNA/PEI, (D) siRNA/PEI/YY-DNAs, (E) siRNA/PLL and (F) siRNA/PLL/YY-DNAs. | ||
Second, the electrostatic encapsulation of siRNA/LP, siRNA/PEI and siRNA/PLL binary complexes with dendrimer-like polymeric DNAs (YY-DNAs) was performed at varying weight ratio of YY-DNAs, and the complex formation was confirmed by a gel retardation assay (Fig. 2B, D and F). As a result, the bands of siRNA and YY-DNAs were not detected at weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and 1
4 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, while a slight bands of YY-DNAs were observed at a weight ratio of 1
6, while a slight bands of YY-DNAs were observed at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8. These results indicate that the binary complexes could be encapsulated by YY-DNAs without releasing of siRNA, and formed stable anionic complexes. In addition, the maximum encapsulation capacity of binary complex was 1
8. These results indicate that the binary complexes could be encapsulated by YY-DNAs without releasing of siRNA, and formed stable anionic complexes. In addition, the maximum encapsulation capacity of binary complex was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 weight ratio.
6 weight ratio.
Recently, the morphology of siRNA/cationic vectors such as siRNA/PEI24 and pDNA/PEI coated with anionic polylactic acid25 were investigated, which showed spherical shapes. On the same manner, scanning electron microscopy was used to examine the morphology of the cationic binary complexes of siRNA/LP, siRNA/PEI and siRNA/PLL at weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 and the anionic encapsulated complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at weight ratio of 1
8 and the anionic encapsulated complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (Fig. 3). Both cationic and anionic complexes showed nearly spherical morphology.
4 (Fig. 3). Both cationic and anionic complexes showed nearly spherical morphology.
Generally, cellular uptake and distribution of complexes depend on the size of complexes. Sakurai et al. reported that nanoparticles smaller than 200 nm were observed in retinal cells other that the vitreous cavity and trabecular meshwork where only lager diameter particles were distrusted after intravitreal injection.26 Thus, small particle size is an important factor for effective siRNA delivery. In order to investigate the effect of encapsulation on the particle size, dynamic light scattering was used to determine the hydrodynamic diameter and polydisperstiy index of anionic encapsulated siRNA complexes as shown in Table 1. The binary complexes of siRNA/LP, siRNA/PEI and siRNA/PLL showed 120, 86.6 and 65.4 nm hydrodynamic diameters and 0.26, 0.21 and 0.18 polydisperstiy index, respectively. While the encapsulation of these binary complexes with anionic YY-DNAs causes a significant decreasing in the hydrodynamic diameter. The decreasing in the size of the anionic encapsulated complexes with the increasing of YY-DNAs ratio upon weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 may be attributed to shrinkage of the cationic vectors through the strong electrostatic interaction with YY-DNAs.
6 may be attributed to shrinkage of the cationic vectors through the strong electrostatic interaction with YY-DNAs.
| Complex | Size (hydrodynamic diameter, nm) | Polydisperstiy index | ζ-potential (mV) |
|---|---|---|---|
| a Each data represents the mean ± S.D. (n = 3). | |||
siRNA/LP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8) 8) |
120 ± 12.3 | 0.26 ± 0.08 | +65.5 ± 2.3 |
siRNA/LP/YY-DNAs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
40.21 ± 9.3 | 0.17 ± 0.07 | −8.6 ± 3.6 |
siRNA/LP/YY-DNAs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
37.9 ± 4.2 | 0.16 ± 0.03 | −17.3 ± 2.4 |
siRNA/LP/YY-DNAs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) 6) |
64.4 ± 18.0 | 0.19 ± 0.14 | −19.6 ± 3.2 |
siRNA/PEI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8) 8) |
86.6 ± 14.5 | 0.21 ± 0.09 | +45.6 ± 1.2 |
siRNA/PEI/YY-DNAs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
15 ± 6.6 | 0.13 ± 0.05 | −22.4 ± 2.8 |
siRNA/PEI/YY-DNAs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
13 ± 9.6 | 0.11 ± 0.07 | −28.8 ± 0.9 |
siRNA/PEI/YY-DNAs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) 6) |
15.1 ± 5.9 | 0.13 ± 0.05 | −26.5 ± 1.5 |
siRNA/PLL (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8) 8) |
65.4 ± 16.8 | 0.18 ± 0.10 | +32.1 ± 1.1 |
siRNA/PLL/YY-DNAs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
15.16 ± 6.8 | 0.13 ± 0.06 | −23.3 ± 0.8 |
siRNA/PLL/YY-DNAs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) 4) |
11.99 ± 3.7 | 0.12 ± 0.04 | −29.6 ± 1.4 |
siRNA/PLL/YY-DNAs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) 6) |
18.95 ± 3.2 | 0.14 ± 0.03 | −27.8 ± 2.6 |
On the other hand, the ζ-potential of siRNA complexes were determined and shown in Table 1. The addition of YY-DNAs to the positively charged siRNA binary complexes gave negative ζ-potential values, suggesting the encapsulation of binary siRNA complexes with anionic YY-DNAs.
3.3. In vitro release of siRNA from the anionic encapsulated complexes
Fluorescence spectroscopy was used to study the release profile of FAM-siRNA from the anionic encapsulated siRNA complexes in vitro. The anionic complexes containing 5 μg of FAM-siRNA were incubated at 37 °C in 1× PBS, pH 7.4 for different times. As shown in Fig. 4, a low initial burst of 15–25% of the anionic complexes was observed for the first 1–3 days followed by continuous release of siRNA. Then the total percent of encapsulated siRNA released from anionic complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs were ∼45%, ∼60% and ∼55% in 7 days, respectively.3.4. Cytotoxicity assay of anionic encapsulated siRNA complexes
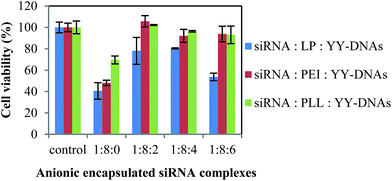
The reduced cytotoxicity after prolonged incubation with synthetic vectors might be one of the main criteria for successful in vivo transfection. Therefore, we evaluated the cytotoxicity of our developed anionic encapsulated siRNA complexes with the cell counting kit-8 (CCK-8) assay after 24 h incubation in HeLa cells, as shown in Fig. 5. The siRNA/LP, siRNA/PEI and siRNA/PLL complexes showed extremely high cytotoxicity. The strong positive charge of the cationic complexes was reported to cause a strong interaction with negatively charged cell membrane and thus causes cell death and cytotoxicity.27 On contrast, all anionic complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at varying weight ratios, did not show significant cytotoxicity. Since, these encapsulated siRNA complexes with a negative ζ-potential value might hardly interact with the negatively charged cell membranes, loading to reduce cytotoxicity in long-term incubation.3.5. Interaction with erythrocytes
For safe siRNA delivery in vivo, the siRNA complexes must be diffused through the vascular before they reached the target cells without aggregation with the erythrocytes. Therefore, the aggregation of erythrocytes with anionic encapsulated siRNA complexes and the turbidity of erythrocytes mixed with theses complexes were determined to evaluate the suitability of the complexes for intraocular injection. Cationic siRNA delivery vectors such as PEI, lead to the aggregation of erythrocytes by the strong affinity of positively charge vector to the cellular membrane.8,28 In this experiment, cationic siRNA/LP, siRNA/PEI and siRNA/PLL complexes at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 also showed severe aggregation with erythrocytes while, no aggregation was observed with the anionic encapsulated siRNA complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at a weight ratio of 1
8 also showed severe aggregation with erythrocytes while, no aggregation was observed with the anionic encapsulated siRNA complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (Fig. 6). Furthermore, Fig. 7 shows the turbidity of the erythrocytes with various siRNA complexes. The cationic siRNA complexes increased the turbidity of erythrocytes. In contrast, all anionic siRNA encapsulated complexes showed much low turbidity. These results indicated that the negatively charged surface of the encapsulated siRNA complexes could prevent the aggregation and turbidity of erythrocytes and thus our anionic complexes might be applied to safe siRNA delivery in clinical applications.
4 (Fig. 6). Furthermore, Fig. 7 shows the turbidity of the erythrocytes with various siRNA complexes. The cationic siRNA complexes increased the turbidity of erythrocytes. In contrast, all anionic siRNA encapsulated complexes showed much low turbidity. These results indicated that the negatively charged surface of the encapsulated siRNA complexes could prevent the aggregation and turbidity of erythrocytes and thus our anionic complexes might be applied to safe siRNA delivery in clinical applications.
3.6. Cellular uptake of anionic encapsulated siRNA complexes
To evaluate the potential of the anionic siRNA encapsulated complexes for effective siRNA delivery and comparing them with the binary siRNA complexes, in vitro cellular uptake experiments were performed on HeLa cells. Binary complexes of siRNA/LP, siRNA/PEI and siRNA/PLL at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 and anionic encapsulated complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at a weight ratio of 1
8 and anionic encapsulated complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 containing 1 μM of FITC-labeled siRNA were transfected into HeLa cells for 4 h and then the cells were observed by fluorescence microscopy. As shown in Fig. 8, siRNA was not observed in the cells. While, the binary complexes of siRNA/LP, siRNA/PEI and siRNA/PLL and all anionic encapsulated siRNA complexes were highly observed into the cells. The results showed that approximately 60–80% of HeLa cells have been transfected with both binary siRNA and anionic encapsulated siRNA complexes. The cationic siRNA/PEI or pDNA/PEI complex was reported to be taken by cells through endocytosis according to electrostatic interaction with cell membrane,29 thus, anionic complex could not be taken by cells in the same manner. Amazing, the anionic encapsulated siRNA complexes were highly taken up by the cells. These results indicate that the specific mechanism participates in the uptake of the siRNA complex with a negative charge. Recently, pDNA/PEI coated with anionic γ-PGA polymer8 or CS30 was reported to be taken by the cells through an energy-dependent process and γ-PGA-/CS-specific receptor-mediated pathway. In the present study, the anionic complex might be recognized by a receptor and taken by the cells via receptor-mediated endocytosis and released siRNA to cytoplasm from endosome by proton sponge mechanisms of PEI; and then the complex showed high gene expressions. Further study should be performed.
4 containing 1 μM of FITC-labeled siRNA were transfected into HeLa cells for 4 h and then the cells were observed by fluorescence microscopy. As shown in Fig. 8, siRNA was not observed in the cells. While, the binary complexes of siRNA/LP, siRNA/PEI and siRNA/PLL and all anionic encapsulated siRNA complexes were highly observed into the cells. The results showed that approximately 60–80% of HeLa cells have been transfected with both binary siRNA and anionic encapsulated siRNA complexes. The cationic siRNA/PEI or pDNA/PEI complex was reported to be taken by cells through endocytosis according to electrostatic interaction with cell membrane,29 thus, anionic complex could not be taken by cells in the same manner. Amazing, the anionic encapsulated siRNA complexes were highly taken up by the cells. These results indicate that the specific mechanism participates in the uptake of the siRNA complex with a negative charge. Recently, pDNA/PEI coated with anionic γ-PGA polymer8 or CS30 was reported to be taken by the cells through an energy-dependent process and γ-PGA-/CS-specific receptor-mediated pathway. In the present study, the anionic complex might be recognized by a receptor and taken by the cells via receptor-mediated endocytosis and released siRNA to cytoplasm from endosome by proton sponge mechanisms of PEI; and then the complex showed high gene expressions. Further study should be performed.
For a more quantitative analysis of the uptake efficiency of siRNA complexes, the fluorescence intensity of the cell lysate of the transfected cells with FITC-labeled siRNA complexes for 4 h into 0.5 mL eppendorf tubes at 37 °C was measured by using a FP-8200 spectrofluorometer (Fig. 9). The uptake efficiency of anionic encapsulated siRNA complexes was comparable to that of cationic siRNA complexes. As example, the uptake efficiencies in HeLa cells treated with the anionic encapsulated complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 were 60.77 ± 0.84, 24.01 ± 2.41 and 40.85 ± 14.12 nM of the intracellular concentration of siRNA (Fig. 9A), 77.59 ± 27.57, 15 ± 0.85 and 23.19 ± 7.00 transmittance per 1 mg mL−1 protein (Fig. 9B) and 0.51 ± 0.01, 0.31 ± 0.02 and 0.38 ± 0.1 transmittance per 1 HeLa cell (Fig. 9C) which approximately similar to that of cationic complexes of siRNA/LP, siRNA/PEI and siRNA/PLL, respectively. These results indicated that the anionic siRNA encapsulated complexes possess a highly ability to taken by HeLa cells even they had negative charge. Therefore, these newly complexes might be used for safe and effective delivery of siRNA in many gene therapy assays.
4 were 60.77 ± 0.84, 24.01 ± 2.41 and 40.85 ± 14.12 nM of the intracellular concentration of siRNA (Fig. 9A), 77.59 ± 27.57, 15 ± 0.85 and 23.19 ± 7.00 transmittance per 1 mg mL−1 protein (Fig. 9B) and 0.51 ± 0.01, 0.31 ± 0.02 and 0.38 ± 0.1 transmittance per 1 HeLa cell (Fig. 9C) which approximately similar to that of cationic complexes of siRNA/LP, siRNA/PEI and siRNA/PLL, respectively. These results indicated that the anionic siRNA encapsulated complexes possess a highly ability to taken by HeLa cells even they had negative charge. Therefore, these newly complexes might be used for safe and effective delivery of siRNA in many gene therapy assays.
The biodistribution of high negatively charged siRNA delivery systems after intravenous injection into mice was recently reported.15 Harrorti et al., injected anionic polymer-coated lipoplexes with CS, PGA and poly-aspartic acid (PAA) of cy5.5 siRNA into mice and observed the biodistribution of siRNA at 1 h after the injection by fluorescence microscopy. It was found that the anionic CS-, PGA- and PAA coated lipoplexes were largely accumulated in the liver and the kidneys. These results inducted that these anionic complexes have a good biodistribution and potential as a targeting vector of siRNA to the liver. Therefore, our newly anionic complexes might be have a good biodistribution after injection into mice and be suitable for the in vivo applications.
3.7. Gene silencing effect
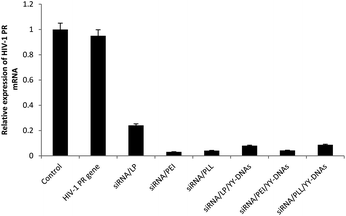
To clearly confirm the cellular uptake of anionic encapsulated siRNA complexes and whether the encapsulated siRNA can induced RNAi in the cells as expected, we evaluated the silencing effect of the anionic complexes on the expression of HIV-PR gene with HeLa cells. Binary complexes of siRNA/LP, siRNA/PEI and siRNA/PLL at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 and anionic siRNA encapsulated complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at a weight ratio of 1
8 and anionic siRNA encapsulated complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs at a weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 containing 1.5 μg of HIV-1 PR siRNA were evaluated into HeLa cells. As expected, all cationic binary complexes of siRNA/LP, siRNA/PEI and siRNA/PLL exhibited approximately 76%, 96% and 96% of HIV-PR gene silencing, respectively (Fig. 10). In addition, all anionic complexes knocked down HIV-PR mRNA expression (Fig. 10). We found that siRNA transfected by our anionic encapsulated complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs achieved approximately 92%, 96% and 92% gene silencing efficiencies, which more than that of the commercially available transfection reagent (LP) and comparable with other two reported (PEI and PLL). These results indicated that the anionic siRNA encapsulated complexes possess a highly ability to taken by HeLa cells even they had negative charge and the encapsulated siRNA has extremely high gene silencing effect.
4 containing 1.5 μg of HIV-1 PR siRNA were evaluated into HeLa cells. As expected, all cationic binary complexes of siRNA/LP, siRNA/PEI and siRNA/PLL exhibited approximately 76%, 96% and 96% of HIV-PR gene silencing, respectively (Fig. 10). In addition, all anionic complexes knocked down HIV-PR mRNA expression (Fig. 10). We found that siRNA transfected by our anionic encapsulated complexes of siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs achieved approximately 92%, 96% and 92% gene silencing efficiencies, which more than that of the commercially available transfection reagent (LP) and comparable with other two reported (PEI and PLL). These results indicated that the anionic siRNA encapsulated complexes possess a highly ability to taken by HeLa cells even they had negative charge and the encapsulated siRNA has extremely high gene silencing effect.
It was reported that lipid-like (lipidoid) nanoparticles such as epoxide-derived lipidoids31 and yne-derived lipidoids32 were used to deliver siRNA in vitro. Among fifty epoxide-derived lipidoids, only three lipidoids exhibited gene silencing efficiencies of 90%. While, among thirty three yne-derived lipidoids, only 40% of them could be silenced greater than 50%. Therefore, our anionic encapsulated complexes were improved the gene silencing efficiency over the reported ones. Furthermore, all these lipidoids are chemically composed materials which might have high cytotoxicity on cells at high concentrations. On contrast, our anionic encapsulated complexes didn't show any toxicity on cells even at high concentration 2 μg of siRNA. Therefore, these newly complexes might be used for safe and effective delivery of siRNA in many gene therapy assays.
3.8. Stability study
The serum stability of siRNA complexes is important for in vivo applications. Therefore, we investigated the stability of anionic siRNA encapsulated complex of siRNA/PLL/YY-DNAs in 10% fetal bovine serum (FBS) and human serum. siRNA/PLL/YY-DNAs complex containing 1.9 μM of siRNA was incubated at 37 °C in 10% FBS or human serum for different times. Its degradation was analyzed by agarose gel electrophoresis visualizing with ethidium bromide. Corresponding YY-DNAs and siRNA in 1× PBS, pH 7.4 were served as control. As shown in Fig. 11A, siRNA/PLL/YY-DNAs complex showed no obvious signs of degradation in 10% FBS even at 72 h, since no detectable siRNA or YY-DNAs band was seen. Furthermore, the incubation of the complex in human serum showed high stability until 48 h. After 48 h, almost of anionic siRNA complex was degraded into many several fragments (Fig. 11B).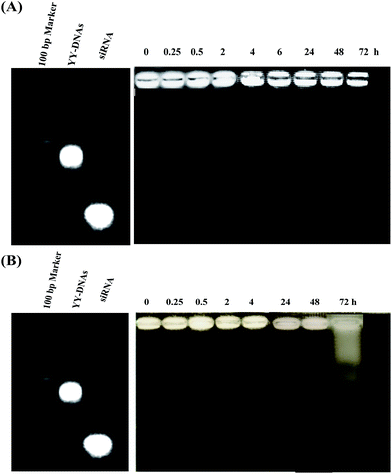 | ||
| Fig. 11 Serum stability of siRNA/PLL/YY-DNAs complex in (A) 10% fetal bovine serum and (B) human serum. | ||
4. Conclusions
In this study, we developed a safe and efficient siRNA delivery system based on the electrostatic encapsulation of siRNA/cationic vectors complexes with YY-DNAs to form stable anionic complexes of siRNA/cationic vector/YY-DNAs. Compared with the cationic complexes of siRNA/LP siRNA/PEI and siRNA/PLL, the advantages of our anionic siRNA encapsulated complexes are that the encapsulation of YY-DNAs of these cationic siRNA complexes dramatically decreased the cytotoxicity on HeLa cell membranes and agglutination with erythrocytes. Furthermore, these anionic siRNA encapsulated complexes were highly taken by the HeLa cells via a receptor-mediated pathway and showed extremely high cellular uptake efficiencies and gene silencing effects even they had negative charge, which more than that of the commercially available transfection reagent (LP). Further studies are necessary to examine the detailed uptake mechanism.Among siRNA/LP/YY-DNAs, siRNA/PEI/YY-DNAs and siRNA/PLL/YY-DNAs complexes, we discovered that the siRNA/PEI/YY-DNAs at weight ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 showed higher gene expression without cytotoxicity and agglutination of erythrocytes. The stability of these anionic siRNA complexes in 10% FBS and human serum was investigated and they showed high stability even after incubation for 72 h and 48 h, respectively. Therefore, the anionic siRNA encapsulated complexes will be very useful for the gene therapy and further experiments may be necessary to investigate their clinical usefulness.
4 showed higher gene expression without cytotoxicity and agglutination of erythrocytes. The stability of these anionic siRNA complexes in 10% FBS and human serum was investigated and they showed high stability even after incubation for 72 h and 48 h, respectively. Therefore, the anionic siRNA encapsulated complexes will be very useful for the gene therapy and further experiments may be necessary to investigate their clinical usefulness.
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and also supported partially by the Global Center of Excellence Program at Nagasaki University.Notes and references
- A. Reynolds, D. Leake, Q. Boese, S. Scaringe, W. S. Marshall and A. Khvorova, Nat. Biotechnol., 2004, 22, 326–330 CrossRef CAS PubMed.
- S. J. Tebes and P. A. Kruk, Gynecol. Oncol., 2005, 99, 736–741 CrossRef CAS PubMed.
- W. Jiangyu, H. Weizhe and H. Ziying, Sci. World J., 2013, 13, 1–16 Search PubMed.
- B. R. Meade and S. F. Bowdy, Adv. Drug Delivery Rev., 2007, 59, 134–140 CrossRef CAS PubMed.
- J. Wang, Z. Lu and M. G. Wientjes, AAPS J., 2010, 12, 492–503 CrossRef CAS PubMed.
- H. Eliyahu, N. Servel, A. J. Domb and Y. Barenholz, Gene Ther., 2002, 9, 850–858 CrossRef CAS PubMed.
- D. Simberg, S. Weisman, Y. Talmon, A. Faerman, T. Shoshani and Y. Barenholz, J. Biol. Chem., 2003, 278, 39858–39865 CrossRef CAS PubMed.
- T. Kurosaki, T. Kitahara, S. Fumoto, K. Nishida, J. Nakamura, T. Niidome, Y. Kodama, H. Nakagawa, H. To and H. Sasaki, Biomaterials, 2009, 30, 2846–2853 CrossRef CAS PubMed.
- E. J. Oh, K. Park and K. S. Kim, J. Controlled Release, 2010, 141, 2–12 CrossRef CAS PubMed.
- T. Kurosaki, T. Kitahara, S. Fumoto, K. Nisida, K. Yamamoto and H. Nakagawa, J. Pharm. Pharm. Sci., 2010, 13, 351–361 CAS.
- N. Duceppe and M. Tabrizian, Biomaterials, 2009, 30, 2625–2631 CrossRef CAS PubMed.
- T. Ito, Y. Koyama and M. Otsuka, J. Pharm. Biomed. Anal., 2010, 51, 268–272 CrossRef CAS PubMed.
- N. Nishiyama, A. Iriyama, W. Jang, K. Miyata, K. Itaka, Y. Inoue, H. Takahashi, Y. Yanagi, Y. Tamaki, H. Koyama and K. Kataoka, Nat. Mater., 2005, 4, 934–941 CrossRef CAS PubMed.
- Y. Kodama, S. Harauchi, S. Kawanabe, N. Ichikawa, H. Nakagawa, T. Muro, N. Higuchi, T. Nakamura, T. Kitahara and H. Sasaki, Biol. Pharm. Bull., 2013, 36, 995–1001 CAS.
- H. Yoshiyuki, N. Ayako, A. Shohei, N. Mayu, O. Hiroyuki, K. Kumi, Y. Maitani and E. Yonemochi, Results Pharma Sci., 2014, 4, 1–7 CrossRef PubMed.
- A. F. M. EL-Mahdy, T. Shibata, T. Kabashima and M. Kai, Chem. Commun., 2014, 50, 859–861 RSC.
- A. F. M. EL-Mahdy, V. Ejupi, T. Shibata, T. Kabashima, J. Lu and M. Kai, Microchim. Acta, 2015, 182, 495–503 CrossRef CAS.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
- Y. Chen, X. Zhu, X. Zhang, B. Liu and L. Huang, Mol. Ther., 2010, 18, 1650–1656 CrossRef CAS PubMed.
- D. W. Bartlett and M. E. Davis, Biotechnol. Bioeng., 2008, 99, 975–985 CrossRef CAS PubMed.
- Q. Ge, L. Filip, A. Bai, T. Nguyen and H. N. Eisen, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8676–8681 CrossRef CAS PubMed.
- B. R. Meade and S. F. Dowdy, Adv. Drug Delivery Rev., 2008, 60, 530–536 CrossRef CAS PubMed.
- J. H. Lee, H. H. Ahn, K. S. Kim, J. Y. Lee, M. S. Kim, B. Lee, G. Khang and H. B. Lee, J. Tissue Eng. Regener. Med., 2008, 2, 288–295 CrossRef CAS PubMed.
- M. Zhang, R. Xu, X. Xia, Y. Yang, J. Gu, G. Qina, X. Liu, M. Ferrari and H. Shen, Biomaterials, 2014, 35, 423–431 CrossRef CAS PubMed.
- H. Laroui, D. Geem, B. Xiao, E. Viennois, P. Rakhya, T. Denning and D. Merlin, Mol. Ther., 2014, 22, 69–80 CrossRef CAS PubMed.
- E. Sakurai, H. Ozeki, N. Kunou and Y. Ogura, Ophthalmic Res., 2001, 33, 31–36 CrossRef CAS PubMed.
- H. Lv, S. Zhang, B. Wang, S. Cui and J. Yan, J. Controlled Release, 2006, 114, 100–109 CrossRef CAS PubMed.
- G. M. Acland, G. D. Aguirre, J. Ray, Q. Zhang, T. S. Aleman, A. V. Cideciyan, S. E. Pearce-Kelling, V. Anand, Y. Zeng, A. M. Maguire, S. G. Jacobson, W. W. Hauswirth and J. Bennett, Nat. Genet., 2001, 28, 92–95 CAS.
- B. Demeneix and J. P. Behr, Adv. Genet., 2005, 53, 217–230 CAS.
- T. Kurosaki, M. Uematsu, K. Shimoda, K. Suzuma, M. Nakai, T. Nakamura, T. Kitahara, T. Kitaoka and H. Sasaki, Biol. Pharm. Bull., 2013, 36, 96–101 CAS.
- K. T. Love, K. P. Mahon, C. G. Levins, K. A. Whitehead, W. Querbes, J. R. Dorkin, J. Qin, W. Cantley, L. L. Qin, T. Racie, M. Frank-Kamenetsky, K. N. Yip, R. Alvarez, D. W. Y. Sah, A. d. Fougerolles, K. Fitzgerald, V. Koteliansky, A. Akinc, R. Langer and D. G. Anderson, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1864–1869 CrossRef CAS PubMed.
- C. A. Alabi, K. T. Love, G. Sahay, H. Yin, K. M. Luly, R. Langer and D. G. Anderson, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12881–12886 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |