Portable and quantitative evaluation of stem cell therapy towards damaged hepatocytes†
Lifang Zhaoa,
Qin Weia,
Hua Wua,
He Li*ab,
Dong Li*c and
Shyam S. Mohapatra*d
aKey Laboratory of Chemical Sensing & Analysis in Universities of Shandong, School of Chemistry and Chemical Engineering, China
bSchool of Biological Science and Technology, University of Jinan, Jinan 250022, China. E-mail: lihecd@gmail.com
cCryomedicine Lab, Qilu Hospital of Shandong University, Jinan 250012, China. E-mail: lidong73@sdu.edu.cn
dDepartment of Internal Medicine, Division of Translational Medicine-Nanomedicine Research Center, Tampa, FL 33612, US. E-mail: smohapat@health.usf.edu
First published on 10th February 2015
Abstract
Stem cell therapy has recently emerged as a breakthrough technology to treat a variety of diseases. Therefore, there is an urgent need to develop appropriate methods to evaluate or monitor stem cell therapy efficiency. Herein, we report a portable and quantitative evaluation method for mesenchymal stem cell (MSC)-based therapy for damaged hepatocytes by ubiquitous personal glucose meters (PGM). It is notable that most current methods for quantitative analysis still require laboratory-based or specialized equipment that is not widely available to the general public. PGMs are one of the devices that are successfully used for in-home medical diagnostics and which have worldwide accessibility to the public. Herein, we report an immunosensor based on PGM for the detection of albumin, the most important indicator for evaluating liver function. Albumin detection can be taken as an appropriate marker to evaluate the efficiency of MSC-based repair of damaged hepatocytes. The proposed sandwich-type immunosensor using PGM exhibits high sensitivity (low detection limit (0.5 ng mL−1), wide range (1 × 10−3 to 10 μg mL−1)), and good reliability. Given the wide availability of antibodies for numerous targets, the proposed method based on PGM can be successfully applied for sensitive detection of many other non-glucose targets, especially helpful for evaluating or monitoring stem cell therapy.
Introduction
Mesenchymal stem cell (MSC)-based cell therapy has emerged as an effective approach to treat a variety of diseases, including acute lung injury,1 kidney injury,2,3 and myocardial infarction.4 Recently, increasing evidence has suggested the therapeutic potential of MSCs in the treatment of liver diseases,5–7 and the transplantation of exogenous MSCs can alleviate acute or chronic liver injury and improve fibrosis.8–10 MSC transfusions exhibited to reverse fulminant hepatic failure and to improve liver function in patients with end-stage liver diseases. In addition to differentiation into functional hepatocytes,11 MSCs can also secrete many cytokines such as interleukin (IL)-10, nerve growth factor, VEGF, HGF, and IGF-1, which have been demonstrated to be effective in regenerating liver structure and promoting recovery of liver function.12–14 Transfusions of umbilical cord-derived MSCs (UC-MSCs) for acute-on-chronic liver failure (ACLF) patients remarkably achieved the increasing survival rates, accompany with increasing serum albumin, cholinesterase, and prothrombin activity. Total bilirubin and alanine aminotransferase levels in serum were obviously decreased with the UC-MSCs transfusions.15 Treatment of liver disease must have an appropriate evaluation system. Apart from morphological observation, apoptosis detection and nucleic acid amplification, methods to assess the effectiveness of stem cell therapy include detecting the secretory function of hepatocytes. Albumin is the major protein secreted by the liver, because it represents ∼50% of the productive effort at any moment and its synthesis amounts to 11–18% of total liver protein synthesis in well-fed animals.16–19 Thus albumin is the most important indicator for evaluating liver function.20 The most commonly used method in detecting secreted proteins is the enzyme linked immunosorbent assay (ELISA), which has the advantage of being quick and convenient. However, the data errors caused by ELISA signal conversion systems have remained a serious shortcoming. Considering the importance of albumin in evaluation of liver function and the requirement for sensitivity and accuracy, it is worthwhile to explore a new system that can detect even small amounts of secreted protein in a multiwell plate cell culture system.As stem cell therapy has emerged as a breakthrough technology in healthcare, developing portable and quantitative methods to evaluate or monitor the therapeutic efficiency becomes much more urgent. It is well known that the accurate detection of specific proteins is playing an increasingly important role in modern healthcare and that the early detection of disease facilitates the application of personalized medicine as well as allowing timely intervention to prevent worsening of the disease.21–24 It is great meaningful to develop portable sensors for fast, reliable and economical detection of target proteins has long been sought, because such sensor will bridge the gap between the wealthy and the poor, as well as between those living in urban area, and those in rural and remote areas with limited access to clinical laboratories.25–27 Notably, the most successful example of such a sensor is the personal glucose meter (PGM).28,29
The PGM was developed over 30 years ago when the first demonstration of using glucose oxidase for glucose monitoring in blood plasma was reported by Clark and Lyons in 1962.30 They have been widely used because of their portability, reliability, accuracy, low cost and easy operation and have either saved or improved the living quality of diabetic patients worldwide.31–33 However, the current PGM can only monitor a single target – blood glucose. Our goal was to find a general method to link the detection of glucose to that of other targets in order to use the PGM as a portable tool for targets other than glucose.34
The aim of this study was to measure the amount of albumin secreted from hepatocytes into the culture supernatant using a commonly available PGM. Here, poly(diallyldimethylammonium chloride) (PDDA)-functionalized graphene oxide (GO) was used to load gold nanoparticles (Au NPs), and then GO–Au NPs were used as a platform for simultaneously immobilizing secondary antibody (Ab2) and invertase. The relationship of various concentrations of albumin and the response signal was obtained, using standard albumin solutions as model analytes. Using the primary antibody (Ab1) immobilized onto Fe3O4–Au NPs as the capture moiety, a rapid and ultrasensitive immunosensor was built and tested to monitor the albumin secreted from hepatocytes into the culture. This strategy was generic and simple and could be applied for portable and quantitative evaluation of stem cell therapy efficiency.
Results and discussion
After several passages, adherent cells from UCs formed a monolayer of typical fibroblastic cells (Fig. S1A†). Flow cytometry results showed that UC-derived cells shared most of their immunophenotype with mesenchymal stem cells, including positive expression for stromal markers (CD29, CD44, CD73, CD90, and CD105) and negative expression for hematopoietic markers (CD34, CD45 and CD133), neural progenitor cell markers CD271, and endothelial cell markers CD31 (Fig. S1B†). These results indicate that we have obtained the cells which are in line with the international criteria of human MSCs.35From preliminary experiments (data not shown), we chose 1 mM H2O2 for 2 hours as appropriate conditions for producing reversible damage to L02. Twenty four hours after hydrogen peroxide treatment, the cellular bodies in the L02 + H2O2 group had expanded and obvious vacuoles had appeared in the cytoplasm. If the medium was replaced with RPMI-1640 containing 20% FBS for 24 hours after injury, the proportion of damaged cells with vacuoles significantly decreased. In addition, the replacement of the medium with CM showed significant improvement of the morphology compared to the L02 + H2O2 group (Fig. 1A–D). Hydrogen peroxide caused a significant acute cell injury as early as 6 hours after treatment. Cells became larger and had a large number of vacuoles in the cytoplasm. Administration of UC-MSC-conditioned medium reduced the ratio of injured L02 cells.
 | ||
| Fig. 1 Conditioned medium from UC-MSCs prevents hydrogen peroxide-induced hepatocyte injury. Error bars represent the standard errors of means (SEM) (n = 3 experiments). | ||
To quantify the effect of hydrogen peroxide and CM on L02 cells, we used an apoptosis assay kit to measure cell apoptosis of each group. As shown in Fig. 1E, the total number of apoptotic cells was significantly higher in the L02 + H2O2 group (24%) than in the normal groups (10.8%) (P < 0.05). Minimal difference in the percentage of early apoptotic cells was found between the L02 + H2O2 group and L02 + H2O2 + CM group (11.1% vs. 10.5%, P > 0.05), whereas a significant difference in the percentage of late apoptotic cells was observed between these two groups (12.9% vs. 6.0%, P < 0.05). Oxidative stress contributes to cellular injury and appears to be the common apoptotic mediator, most likely via lipid peroxidation.36–38 Our study demonstrated that treatment of hepatocytes with H2O2, a precursor of ROS such as highly reactive hydroxyl radicals, leads to cell death via apoptotic processes. Substance that secreted by MSCs enhance cellular antioxidant defences can prevent apoptosis and protect cells from the damaging effects of oxygen radicals. Accumulating evidence has revealed that the therapeutic benefits from MSCs are largely dependent on their capacity to act as a trophic factor pool. After MSCs home to the damaged tissue sites, they will produce a large amount of growth factors performing multiple functions for tissue regeneration.39,40 Many of these factors are critical mediators in angiogenesis and prevention of cell apoptosis, such as vascular endothelial growth factor, insulin-like growth factor 1, basic fibroblast growth factors, hepatocyte growth factor, IL-6, and CCL-2.41,42 In addition, exosomes/microvesicles secreted by MSCs also involved in the process of liver regeneration.43
The morphology of fabricated Fe3O4 beads and Fe3O4–Au nanocomposites was studied by transmission electron microscopy (TEM). The Fe3O4 beads had an average size of ∼70 nm and were uniform in size (Fig. 2A). The ξ-potential of the Fe3O4 spheres was negative at neutral pH due to carboxyl groups on the surface of the Fe3O4 spheres. The ξ-potential of the surface was switched to positive values after the absorption of the positively charged PDDA on the colloidal Fe3O4 beads.44 The prepared Au NPs could be tethered on the surface of PDDA–Fe3O4 nanoparticles because the surface of Au NPs is negatively charged (Fig. 2B). The scanning electron microscopy (SEM) image of the Fe3O4–Au nanocomposites (inset in Fig. 2B) showed that many small Au NPs were adhering to the surface of the PDDA-coated Fe3O4 beads. X-ray diffraction (XRD) was performed to further confirm that Au NPs were successfully assembled onto the surface of Fe3O4 beads, as shown in Fig. 2E. The diffraction peaks of Fe3O4–Au nanocomposites could be indexed to the face-centered cubic structure of magnetite according to JCPDS card no. 65-3107 (circle) and the face-centered cubic structure of gold according to JCPDS card no. 65-8601 (triangle).45 A relatively high amount of Ab1 was conjugated onto the Au NPs through Au–amino intercalations, resulting in a large amount of albumin being easily captured.
Recent research has indicated that GO with abundant oxygen functional groups can be effectively used to load metal nanoparticles and a variety of enzymes.46 As shown in Fig. 2C, the obtained GO was of monolayer flake-like shape which could increase the quantity of immobilization. The GO was negatively charged due to the ionization of functional groups like carboxylic acids located on GO platelets.47 The subsequent absorption of the positively charged PDDA switched the ξ-potential of GO to positive values. After modification with Au NPs, a negatively charged surface of GO–Au was restored, which is due to the adsorption of the negatively charged, citrate-capped Au NPs on the GO surface.48 Fig. 2D shows that the Au NPs distribution and size were uniform, with little aggregation of the GO layers. The energy dispersive X-ray spectroscopy (EDS) analysis was performed to further confirm that the Au NPs successfully adhered to the surface of GO, (Fig. 2F). The characteristic absorption peak of Au verifies the presence of Au NPs on GO. Because of the large amount of Au NPs loaded onto the surface of GO, a large amount of invertase and Ab2 could be easily immobilized onto the GO–Au nanoparticles.
To test the performance of the albumin system, we determined the optimum conditions by quantitative detection of glucose using PGM. Fig. S2–S6† shows the PGM response during optimization with various influencing factors. In order to detect trace amounts of albumin, a 45 °C reaction temperature, an operating pH of 7.0, a 30 minute reaction time with sucrose, 1.5 mg mL−1 invertase and 0.5 M final sucrose concentration were chosen to give the highest signals for the standard procedure. The invertase immobilized on GO–Au–Ab2@invertase bioconjugates converted sucrose into glucose whose concentration could be read using the PGM and albumin detected as a result.
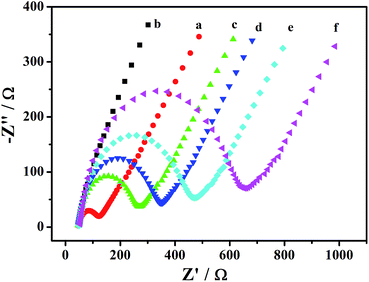
Furthermore, electrochemical impedance spectroscopy (EIS) was performed to give information on the impedance change of the electrode surface in the modification process. In the EIS, the linear portion at low frequencies is associated with electrochemical behavior limited by diffusion. The semicircle portion at high frequencies is, however, related to the electrochemical process subject to electronic transfer, where the diameter corresponds to the electron-transferring resistance (Ret). Considering the stepwise assembly and coating involved in the immunosensor, the fabrication in each step was elucidated by a comparative study. As shown in Fig. 3, the glassy-carbon electrode (GCE; curve a) appeared to be linear with less semicircle, while the Fe3O4–Au-modified electrode (curve b) appeared to be linear with no semicircle; this indicated that the Fe3O4–Au-favored acceleration of the electron transfer was successfully modified onto GCE. After the electrode was coated with Fe3O4–Au–Ab1, the Ret increased a little (Fig. 3c), indicating that Ab1 was successfully attached to the surface of Fe3O4–Au. Coating the electrode with BSA and albumin resulted in the obvious reinforcement of these semicircle portions (curves d and e); and the reinforcement became more evident with further incubation of GO–Au–Ab2@invertase (curve f). These results were consistent with the conclusion that the protein layer insulated the conductive support and perturbed the interface. Therefore, it was clear that the assembly of these biomolecules on the surface of the electrode to form an immunosensor was successful and that the use of a PGM-based immunosensor for the evaluation of stem cell therapy efficiency was feasible.
The standard calibration curve for albumin detection is shown in Fig. 4A. The PGM reading increased with the increase of albumin concentration in the range from 1 × 10−3 to 10 μg mL−1, with a detection limit of 0.5 ng mL−1. The calibration curve was linear with a correlation coefficient of R2 = 0.995 (Y = 5.112![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c + 20.045) in the range of 1 × 10−3 to 10 μg mL−1 (S/N = 3). The low detection limit may be attributed to two aspects: (1) the high loading level of Au NPs on Fe3O4 increased the amount of Ab1 conjugated onto Fe3O4 and (2) the large amount of invertase immobilized onto GO–Au nanoparticles enhanced the ability to convert sucrose to glucose, the original signal source for the PGM. Thus, even trace amounts of albumin can generate a readable signal with the PGM.
c + 20.045) in the range of 1 × 10−3 to 10 μg mL−1 (S/N = 3). The low detection limit may be attributed to two aspects: (1) the high loading level of Au NPs on Fe3O4 increased the amount of Ab1 conjugated onto Fe3O4 and (2) the large amount of invertase immobilized onto GO–Au nanoparticles enhanced the ability to convert sucrose to glucose, the original signal source for the PGM. Thus, even trace amounts of albumin can generate a readable signal with the PGM.
 | ||
| Fig. 4 (A) Calibration curve of the immunosensor toward different concentrations of albumin. Error bar = RSD (n = 3) (B) comparison of albumin concentrations detected by PGM method with ELISA. | ||
To evaluate the analytical reliability and application potential of the proposed method for clinical evaluation of albumin using a PGM, the assay results of cultural supernatants using the PGM were compared with the results by ELISA. The albumin concentration determined by the PGM method fit well with the ELISA results. The data obtained by the two ways give a straight line with a correlation coefficient of 0.993 (Fig. 4B), indicating a good likelihood for application of the PGM method in the clinic.
For evaluation of MSC-based therapy towards damaged hepatocytes, cultural supernatants were detected using the PGM method. When the levels of albumin in serum samples were over the calibration range, serum samples were diluted with PBS. The results of four types albumins secreted from hepatocytes into the culture supernatant are shown in Fig. 5. The results show that supernatants from undamaged L02 liver cells with a typical polygonal morphology (Fig. 1A) have the highest albumin concentration. After hydrogen peroxide damage, cytoplasmic vacuoles began to appear and there was a dramatic fall in supernatant concentration of albumin. Some cells showed severe contraction and disintegration (Fig. 1B). The cells treated with RPMI-1640 medium had a restored level of albumin in supernatant. Especially, the group receiving MSC-conditioned medium had higher levels of albumin than the RPMI-1640 medium treatment group, which indicated that MSCs can repair damaged synthetic function of hepatocytes. The results by PGM fit well with the observation that UC-MSC-conditioned medium prevents hydrogen peroxide-induced hepatocyte injury (Fig. 1). Hence, the PGM immunoassay methodology developed here could be satisfactorily applied to evaluate or monitor the effectiveness of MSC-based therapy towards damaged hepatocytes.
Conclusions
In summary, we have demonstrated a portable, low-cost and quantitative device employing PGM as the signal transducer for detecting albumin to evaluate MSC-based therapy for damaged hepatocytes. By using GO–Au NPs loaded with multiple invertase molecules as signal amplifiers, a low level of albumin of 0.5 ng mL−1 could be unambiguously detected. Due to the cost-effective, easy-to-operate, and commercial availability of PGM to the public, the methodology here could be applied for portable and quantitative evaluation of many non-glucose analytes.Acknowledgements
The authors would like to thank the National Science Foundation of China (no. 21245007, 81000976 and 21075052), the Natural Science Foundation of Shandong Province (no. ZR2010BQ010) and QW thanks the Special Foundation for Taishan Scholar Professorship of Shandong Province and UJN (no. ts20130937) for financial support.Notes and references
- J. Li, D. Li, X. Liu, S. Tang and F. Wei, J. Inflammation, 2012, 9, 33 CrossRef CAS PubMed.
- Y. Chen, H. Qian, W. Zhu, X. Zhang, Y. Yan, S. Ye, X. Peng, W. Li and W. Xu, Stem Cells Dev., 2010, 20, 103 CrossRef PubMed.
- H. Cao, H. Qian, W. Xu, W. Zhu, X. Zhang, Y. Chen, M. Wang, Y. Yan and Y. Xie, Biotechnol. Lett., 2010, 32, 725 CrossRef CAS PubMed.
- M. Hou, K. M. Yang, H. Zhang, W. Q. Zhu, F. J. Duan, H. Wang, Y. H. Song, Y. J. Wei and S. S. Hu, Int. J. Cardiol., 2007, 115, 220 CrossRef PubMed.
- Y. J. Chang, J. W. Liu, P. C. Lin, L. Y. Sun, C. W. Peng, G. H. Luo, T. M. Chen, R. P. Lee, S. Z. Lin and H. J. Harn, Life Sci., 2009, 85, 517 CrossRef CAS PubMed.
- Y. Yan, W. Xu, H. Qian, Y. Si, W. Zhu, H. Cao, H. Zhou and F. Mao, Liver Int., 2009, 29, 356 CrossRef CAS PubMed.
- Y. Chen, L. X. Xiang, J. Z. Shao, R. L. Pan, Y. X. Wang, X. J. Dong and G. R. Zhang, J. Cell. Mol. Med., 2010, 14, 1494 CrossRef PubMed.
- S. Oyagi, M. Hirose, M. Kojima, M. Okuyama, M. Kawase, T. Nakamura, H. Ohgushi and K. Yagi, J. Hepatol., 2006, 44, 742 CrossRef CAS PubMed.
- R. Higashiyama, Y. Inagaki, Y. Y. Hong, M. Kushida, S. Nakao, M. Niioka, T. Watanabe, H. Okano, Y. Matsuzaki and G. Shiota, Hepatology, 2007, 45, 213 CrossRef CAS PubMed.
- B. Petersen, W. Bowen, K. Patrene, W. Mars, A. Sullivan, N. Murase, S. Boggs, J. Greenberger and J. Goff, Science, 1999, 284, 1168 CrossRef CAS.
- K. Ishii, Y. Yoshida, Y. Akechi, T. Sakabe, R. Nishio, R. Ikeda, K. Terabayashi, Y. Matsumi, K. Gonda and H. Okamoto, Hepatology, 2008, 48, 597 CrossRef CAS PubMed.
- S. R. Lee, S. H. Lee, J. Y. Moon, J. Y. Park, D. Lee, S. J. Lim, K. H. Jeong, J. K. Park, T. W. Lee and C. G. Ihm, Renal Failure, 2010, 32, 840 CrossRef CAS PubMed.
- N. Lin, K. Hu, S. Chen, S. Xie, Z. Tang, J. Lin and R. Xu, Life Sci., 2009, 85, 291 CrossRef CAS PubMed.
- M. Wang, P. R. Crisostomo, C. Herring, K. K. Meldrum and D. R. Meldrum, Am. J. Physiol. Regul. Integr. Comp. Physiol., 2006, 291, R880–R884 CrossRef CAS PubMed.
- M. Shi, Z. Zhang, R. Xu, H. Lin, J. Fu, Z. Zou, A. Zhang, J. Shi, L. Chen and S. Lv, Stem Cells Transl. Med., 2012, 1, 725 CrossRef CAS PubMed.
- T. Peters, Adv. Protein Chem., 1985, 37, 161 CrossRef CAS.
- T. Peters and J. C. Peters, J. Biol. Chem., 1972, 247, 3858 CAS.
- G. Schreiber and J. Urban, Rev. Physiol. Biochem. Pharmacol., 1978, 82, 27 CAS.
- C. H. van den Akker, H. Schierbeek, T. Rietveld, A. Vermes, J. J. Duvekot, E. A. Steegers and J. B. van Goudoever, Am. J. Clin. Nutr., 2008, 88, 997 CAS.
- H. M. Rotermund, G. Schreiber, H. Maeno, U. Weinssen and K. Weigand, Cancer Res., 1970, 30, 2139 CAS.
- Y. Xiang and Y. Lu, Nat. Chem., 2011, 3, 697 CrossRef CAS PubMed.
- A. S. Daar, H. Thorsteinsdóttir, D. K. Martin, A. C. Smith, S. Nast and P. A. Singer, Nat. Genet., 2002, 32, 229 CrossRef CAS PubMed.
- M. Yang, J. Jiang, Y. Lu, Y. He, G. Shen and R. Yu, Biomaterials, 2007, 28, 3408 CrossRef CAS PubMed.
- Q. Xie, Y. Zhao, X. Chen, H. Liu, D. G. Evans and W. Yang, Biomaterials, 2011, 32, 6588 CrossRef CAS PubMed.
- Y. Xiang and Y. Lu, Anal. Chem., 2012, 84, 4174 CrossRef CAS PubMed.
- Y. Ren, H. Deng, W. Shen and Z. Gao, Anal. Chem., 2013, 85, 4784 CrossRef CAS PubMed.
- Z. Chen, Y. Liu, Y. Wang, X. Zhao and J. Li, Anal. Chem., 2013, 85, 4431 CrossRef CAS PubMed.
- Z. Nie, F. Deiss, X. Liu, O. Akbulut and G. M. Whitesides, Lab Chip, 2010, 10, 3163 RSC.
- P. Heinzerling, F. Schrader and S. Schanze, J. Chem. Educ., 2012, 89, 1582 CAS.
- L. C. Clark and C. Lyons, Ann. N. Y. Acad. Sci., 1962, 102, 29 CrossRef CAS PubMed.
- M. Montagnana, M. Caputo, D. Giavarina and G. Lippi, Clin. Chim. Acta, 2009, 402, 7 CrossRef CAS PubMed.
- J. Su, J. Xu, Y. Chen, Y. Xiang, R. Yuan and Y. Chai, Chem. Commun., 2012, 48, 6909 RSC.
- J. Xu, B. Jiang, J. Xie, Y. Xiang, R. Yuan and Y. Chai, Chem. Commun., 2012, 48, 10733 RSC.
- L. Yan, Z. Zhu, Y. Zou, Y. Huang, D. Liu, S. Jia, D. Xu, M. Wu, Y. Zhou and S. Zhou, J. Am. Chem. Soc., 2013, 135, 3748 CrossRef CAS PubMed.
- M. Dominici, K. Le Blanc, I. Mueller, I. Slaper-Cortenbach, F. Marini, D. Krause, R. Deans, A. Keating, D. Prockop and E. Horwitz, The International Society for Cellular Therapy Position Statement, Cytotherapy, 2006, 8, 315 CrossRef CAS PubMed.
- T. M. Buttke and P. A. Sandstrom, Immunol. Today, 1994, 15, 7 CrossRef CAS.
- Y. J. Choi, J.-S. Kang, J. H. Y. Park, Y. J. Lee, J. S. Choi and Y. H. Kang, J. Nutr., 2003, 133, 985 CAS.
- J. Spencer, H. Schroeter, G. Kuhnle, S. Srai, R. Tyrrell, U. Hahn and C. Rice-Evans, Biochem. J., 2001, 354, 493 CrossRef CAS.
- P. R. Crisostomo, Y. Wang, T. A. Markel, M. Wang, T. Lahm and D. R. Meldrum, Am. J. Physiol.: Cell Physiol., 2008, 294, C675 CrossRef CAS PubMed.
- A. I. Caplan and J. E. Dennis, J. Cell. Biochem., 2006, 98, 1076 CrossRef CAS PubMed.
- G. Xu, Y. Zhang, L. Zhang, G. Ren and Y. Shi, Biochem. Biophys. Res. Commun., 2007, 361, 745 CrossRef CAS PubMed.
- F. Tögel, Z. Hu, K. Weiss, J. Isaac, C. Lange and C. Westenfelder, Am. J. Physiol. Renal. Physiol., 2005, 289, F31 CrossRef PubMed.
- P. Levine, K. McDaniel, H. Francis, L. Kennedy, G. Alpini and F. Meng, Dig. Liver Dis., 2014, 46, 391 CrossRef CAS PubMed.
- T. Zheng, J. J. Fu, L. Hu, F. Qiu, M. Hu, J. J. Zhu, Z. C. Hua and H. Wang, Anal. Chem., 2013, 85, 5609 CrossRef CAS PubMed.
- J. S. Chen, Y. Zhang and X. W. Lou, ACS Appl. Mater. Interfaces, 2011, 3, 3276 CAS.
- R. Martínez-Orozco, H. Rosu, S.-W. Lee and V. Rodríguez-González, J. Hazard. Mater., 2013, 263, 52 CrossRef PubMed.
- B. Li, X. Zhang, X. Li, L. Wang, R. Han, B. Liu, W. Zheng, X. Li and Y. Liu, Chem. Commun., 2010, 46, 3499 RSC.
- Q. Ji, S. Acharya, J. P. Hill, G. J. Richards and K. Ariga, Adv. Mater., 2008, 20, 4027 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra00191a |
| This journal is © The Royal Society of Chemistry 2015 |