Identification of nitric oxide-releasing derivatives of oleanolic acid as potential anti-colon cancer agents†
Junjie Fu‡
ab,
Yu Zou‡a,
Zhangjian Huang*a,
Chang Yana,
Qimeng Zhoua,
Huibin Zhang*a,
Yisheng Laia,
Sixun Penga and
Yihua Zhang*a
aState Key Laboratory of Natural Medicines, Jiangsu Key Laboratory of Drug Discovery for Metabolic Diseases, China Pharmaceutical University, Nanjing 210009, PR China. E-mail: zyhtgd@163.com; cpudahuang@163.com; zhanghb80@163.com
bDepartment of Medicinal Chemistry, College of Pharmacy, University of Florida, Gainesville, FL 32610, USA
First published on 12th February 2015
Abstract
A group of nitric oxide (NO)-releasing derivatives of oleanolic acid (OA) (5–13) were designed, synthesized and biologically evaluated for their anti-colon cancer activity. It was found that the most potent compound 6 preferably released high levels of NO in colon cancer cells but not in normal cells, leading to potent cytotoxicity against colon cancer cells. In addition, 6 was more prone to produce NO catalyzed by GSTπ relative to GSTα. Furthermore, 6 significantly inhibited the tumor growth in a mouse xenograft model. Finally, 6 induced nitration of tyrosine residues in mitochondrial protein, down-regulated Wnt/β-catenin pathway, and inhibited COX-2 centered signaling loop in colon cancer cells. Collectively, our findings suggested that 6 could be a promising candidate for the intervention of colon cancer.
Introduction
Despite extensive research efforts, colon cancer remains the fourth leading cause of cancer death worldwide, affecting men and women of all racial and ethnic groups. Accumulating evidence indicates that colon cancer is caused by alterations of multiple signal pathways due to both genetic and environmental factors, including Wnt/β-catenin, RAS, p53, phosphoinositide-3 kinase (PI3KCA), cyclooxygenase-2 (COX-2)/prostaglandin (PG), etc.1,2 Chemotherapy is one of the most common treatments for colon cancer. However, the application of anticancer agents is often limited by intolerable side effects. Therefore, development of novel molecules with low systematic toxicity and high activity toward colon cancer is in urgent need.There is growing consensus that high levels of nitric oxide (NO) released from NO donors can exert a multitude of anticancer activities, including enhancement of apoptotic stimuli, and inhibition of metastasis, angiogenesis, and hypoxia, thus retarding the cancer cell growth in vitro and in vivo.3–5 The role of NO-releasing drugs in the treatment of colon cancer has also been well documented. For example, NO–aspirin (NO–ASA) inhibited the growth of human colon cell lines HCT-116 and HT-29 by both S-nitrosylation and tyrosine nitration of key signalling proteins such as p53, β-catenin, and NF-κB.6 More recently, it was reported that an NO-releasing derivative of anti-inflammatory drug (S,R)-3-phenyl-4,5-dihydro-5-isoxasole acetic acid (VGX-1027, GIT-27 NO) suppressed the growth of colon cancer cells in vitro and reduced tumor volume in syngeneic BALB/c mice.7
O2-(2,4-Dinitrophenyl)diazeniumdiolate is a known NO donor able to release NO catalyzed by glutathione S-transferase π (GSTπ), which is over-expressed in many human cancer cells8 and is a useful biomarker with greater activity in colon cancer than in adjacent normal tissues.9–12 The characteristic of GSTπ endows compounds containing O2-(2,4-dinitrophenyl)diazeniumdiolate moiety with increased selectivity toward cancer cells. The representative compounds JS-K and PABA/NO (Scheme 1) showed potent anticancer activity in numerous cancer cell lines.13–15 Our group has also developed a number of potent hybrid molecules from O2-(2,4-dinitrophenyl)diazeniumdiolate and oleanolic acid (OA) with selective inhibitory activity against human hepatocellular carcinoma (HCC).16,17 Interestingly, the hybrid 1 (Scheme 1) exhibited improved stability toward glutathione (GSH) in the absence of GSTπ, enhanced water solubility, and better antiproliferative activity against HCC cells as compared with JS-K and PABA/NO.16 Additionally, it is known that OA and its derivatives possess chemotherapeutic and chemopreventive activities against the carcinogenesis and development of colon cancer.18–20 All these findings arise our interest to investigate the inhibitory effects of NO-releasing derivatives of OA on colon cancer. Accordingly, a new pool of these derivatives (5–13) were designed, synthesized and biologically evaluated as potential anti-colon cancer agents.
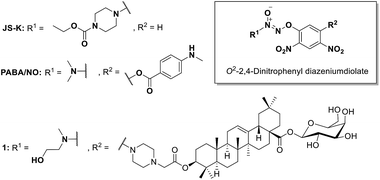 | ||
| Scheme 1 Chemical structures of O2-(2,4-dinitrophenyl)diazeniumdiolate derivatives JS-K, PABA/NO, and diazeniumdiolate–oleanolic acid hybrid 1. | ||
Results and discussion
Rationale for the design of compounds 5–13
Our previous study unambiguously indicates that the anticancer activity of diazeniumdiolate-based NO-releasing derivatives of OA is significantly affected by two moieties. The first, secondary amine moiety to build diazeniumdiolate, and the second, linker moiety between 2,4-dinitrophenyl ring which is attached to diazeniumdiolate, and OA.16 Among the various secondary amines we investigated, N-methylethanolamine (see compound 1, Scheme 1), the only one with a hydroxyl group, was proven to be the optimal for anticancer activity. As for the linker, the best results could be achieved when 2-(piperazin-1-yl)acetic acid was used to link the C-5 position of 2,4-dinitrophenyl to OA (see compound 1, Scheme 1). Based on the above findings, three new secondary amines bearing a hydroxyl group including L-prolinol, 4-hydroxypiperidine, and 1-(2-hydroxylethyl)piperazine (compounds 5–7) were employed in this study. Meanwhile, pyrrolidine and piperidine (compounds 8 and 9) were used as non-hydroxyl counterparts for comparison. On the other hand, 2-(piperazin-1-yl)acetic acid and its two analogs L-proline and N-methylglycine were used as linkers to connect 2,4-dinitrophenyl to OA, respectively (compounds 5–9, and 10–13). In addition, a β-linked galactosyl moiety was attached to C28 of OA to improve the aqueous solubility of the hybrids.Chemistry
The synthetic route for the target compounds 5–13 is depicted in Scheme 2. The corresponding secondary amines were treated with NO gas in the presence of catalytic amount of nanometer-sized TiO2 under atmospheric pressure at room temperature,21 forming diazeniumdiolate sodium salts 2a–f. The O2 anion of the diazeniumdiolate nucleophilically substituted for one of two fluorine atoms in 1,5-difluoro-2,4-dinitrobenzene, yielding O2-(2,4-dinitro-5-fluorophenyl)diazeniumdiolates 3a–f. Finally, substitution of the other fluorine atom by the terminal piperazine nitrogen in 4, which were prepared as previously reported,16 provided target products 5–9. In addition, the 3-OH of 2,3,4,6-tetra-O-acetyl-β-D-galactopyranosyl oleanate (14)22 was coupled with either N-Cbz-protected L-proline or N-Cbz-N-methylglycine in the presence of EDCI and DMAP, affording esters 15a and 15b, respectively. Their acetyl groups in the galactosyl were hydrolyzed using NaOMe to give hydroxyl compounds 16a and 16b. Subsequently, Pd/C-mediated hydrogenation of the Cbz group led to free secondary amines, which were conjugated with O2-(2,4-dinitro-5-fluorophenyl)diazeniumdiolates 3a and 3c to afford target compounds 10–13, respectively.Assessment of in vitro anti-proliferative activity on colon cell lines
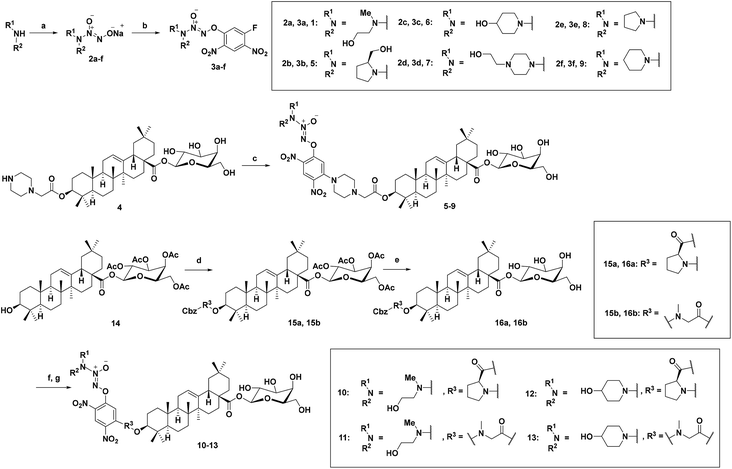
Compounds 5–9 with newly introduced secondary amine in the diazeniumdiolate moiety and 2-(piperazin-1-yl) acetic acid as a linker were firstly examined respectively for their cytotoxicity against three human colon cancer cell lines (HT-29, HCT-15, HCT-116) by MTT assay using JS-K, PABA/NO, and 1 as controls. As shown in Table 1, both 1 and 6 exhibited considerable cytotoxicity on all the three colon cancer cell lines, superior to PABA/NO and comparable with JS-K. Evidently, 6 was the most potent, which inhibited the proliferation of HCT-15 and HCT-116 cells with submicromolar IC50 values (0.924 and 0.823 μM, respectively) (Fig. 1a). Compounds 5 and 7, the other two compounds with a hydroxyl group in the diazeniumdiolate moiety, showed attenuated activity with IC50 values around 10 μM, similar to that of PABA/NO. However, a dramatic decrease in anti-proliferative activity was observed for 8 and 9, which are non-hydroxyl counterparts of 5 and 6, respectively. These results suggest that the hydroxyl group in the diazeniumdiolate moiety might be pivotal for anticancer activity, and 4-hydroxypiperidine be better than N-methylethanolamine. Next, the contribution of linkers L-proline and N-methylglycine to the anticancer activity of 10–13 was evaluated. While all of them exhibited moderate to high activity (Table 1), none of them exceeded 6. Also, 6 showed remarkable inhibitory effects on breast cancer, liver cancer, ovarian cancer, leukemia cell lines, etc. (see ESI†). In sharp contrast, 6 had much less inhibitory activity on normal colon cells CDD-841. It can be seen from Fig. 1b, at 1.25 μM, 6 inhibited the proliferation of HT-29, HCT-15, and HCT-116 cells by 51.4%, 58.5%, and 79.5%, respectively, whereas inhibited that of CDD-841 cells only by 6.33% under the same condition. The cancer cell-selective cytotoxicity of 6 might result from the lower activity of GSTπ in normal cells as compared with cancer cells. Furthermore, pretreatment of HCT-116 cells with GSTπ inhibitor etacrynic acid (EA) significantly diminished the cytotoxicity of 6 (Fig. 1c). These results suggest that GSTπ may be responsible, at least partly, for the anticancer activity of 6.| Compounds | HT-29 | HCT-15 | HCT-116 |
|---|---|---|---|
| a The inhibitory effects of individual compounds on the proliferation of colon cancer cell lines were determined by the MTT assay with a 72 h treatment. Data are expressed as mean ± SD of three independent experiments. | |||
| JS-K | 1.26 ± 0.14 | 0.875 ± 0.07 | 1.49 ± 0.13 |
| PABA/NO | 15.9 ± 1.20 | 6.63 ± 0.72 | 13.2 ± 1.03 |
| 1 | 1.14 ± 0.09 | 2.19 ± 0.18 | 1.40 ± 0.08 |
| 5 | 9.87 ± 0.75 | 8.52 ± 0.90 | 12.3 ± 1.32 |
| 6 | 1.01 ± 0.07 | 0.924 ± 0.05 | 0.803 ± 0.07 |
| 7 | 10.0 ± 1.23 | 13.2 ± 1.14 | 11.3 ± 0.85 |
| 8 | >50 | >50 | 42.3 ± 3.27 |
| 9 | >50 | >50 | >50 |
| 10 | 9.67 ± 0.88 | 7.43 ± 0.56 | 8.05 ± 0.91 |
| 11 | 7.71 ± 1.02 | 5.46 ± 0.36 | 8.32 ± 0.82 |
| 12 | 7.86 ± 0.91 | 7.05 ± 0.55 | 4.89 ± 0.65 |
| 13 | 4.21 ± 0.32 | 6.20 ± 0.74 | 7.01 ± 0.81 |
Assay of intracellular NO release
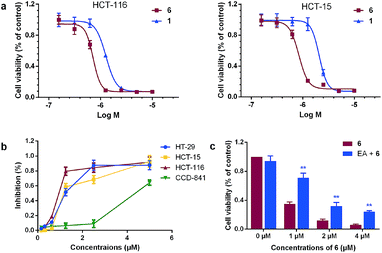
The amounts of NO released in HCT-116 cells by 5–13 were measured using Griess assay. A positive correlationship between the anticancer activity and intracellular NO levels was observed from Fig. 2a and Table 1. Among all the tested target compounds, 6 produced the largest levels of NO in HCT-116 cells, comparable to JS-K and PABA/NO. Noticeable levels of NO production were also detected from active compounds 1 and 12, whereas the two most inactive compounds 8 and 9 produced the lowest levels of NO in the colon cancer cells. Further results showed that 6 concentration- and time-dependently produced high levels of NO in HCT-116 cells but not in normal colon cells CCD-841 (Fig. 2b and c). As expected, pretreatment with GSTπ inhibitor EA or NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (carboxy-PTIO), decreased the NO generation by 6 in HCT-116 cells (Fig. 2d). These results suggest that NO mediated by GSTπ may make an important contribution to the cytotoxicity of 6.Stability evaluation of 6 in the absence of GST
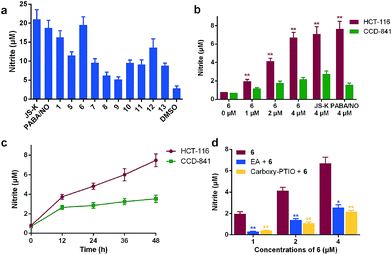
Our previous study has proved that one of the most attracting characteristic of hybrid 1 is its improved resistance toward GSH-induced decomposition in the absence of GSTπ, facilitating the selective accumulation of 1 in cancer cells. This enhanced stability was at least partially attributed to the increased electron density in 2,4-dinitrophenyl ring imparted by the conjugation of OA through a proper linker. Given that the structural difference between 1 and 6 exists only in the diazeniumdiolate part, it is possible that 6 may possess the similar stability like 1. This assumption was verified by examining the decomposition behaviour of 6 in GSH solution. JS-K, PABA/NO, and 6 were incubated in pH 7.4 PBS buffer containing 4 mM GSH at 37 °C, and their reactions with GSH were monitored using HPLC (Fig. 3a). Both JS-K and PABA/NO significantly decomposed within 20 min while most of 6 remained almost intact after 160 min. Compound 6 was next tested in cysteine-rich protein bovine serum albumin (BSA) or homocysteine (Fig. 3b). It was observed that 75.4% and 85.0% of 6 were recovered after a 50 min incubation with BSA and homocysteine, respectively. These results demonstrated that similarly to 1, 6 was much more stable than JS-K and PABA/NO.GSTπ-promoted metabolism of 6
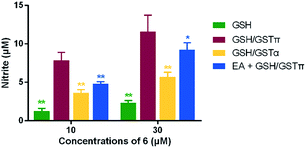
The catalytic effects of GSTπ on the GSH-induced metabolism of 6 was investigated and compared with that of GSTα which predominantly exists in many normal tissues.23 To this end, various concentrations of 6 (10–60 μM) were incubated in pH 7.4 buffer containing 1 mM GSH and a catalytic amount of GSTπ or GSTα (5 μg mL−1). The initial decomposition rate of 6 at each concentration was measured (see ESI†), and kinetic parameters were obtained according to Michaelis–Menten equation. As shown in Table 2, GSTπ catalyzed the metabolism of 6 by GSH with a high catalytic efficiency (18.76 mM−1 s−1), 3.4-fold greater than GSTα (5.53 mM−1 s−1).The NO-releasing capacity of 6 in PBS buffer in the presence of GSH/GSTπ or GSH/GSTα was further studied. Evidently, 6 was more prone to produce NO under the catalysis of GSTπ when compared with that of GSTα, and co-incubation of 6 with GSTπ inhibitor EA partially decreased the NO levels (Fig. 4). Taken together, the metabolism of 6 by GSH to liberate NO was more efficiently catalyzed by GSTπ.
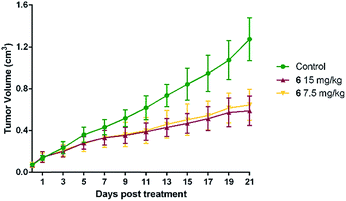
In vivo tumor retarding effects of 6
To evaluate the in vivo anti-colon cancer activity of 6, BALB/c nude mice were inoculated subcutaneously with colon carcinoma cells HCT-116. After the establishment of solid tumor, two groups of the mice were intravenously treated with 7.5 and 15 mg kg−1 of 6, respectively. It could be seen from Fig. 5 that in comparison with the vehicle-treated controls (1.04 ± 0.31 g, P < 0.01, Table 3), treatment with 6 at 15 mg kg−1 significantly inhibited the grow of implanted colon cancer cells, and the tumor weights in the mice were significantly reduced by 60.98%.| Entry | Dose | Tumor volume (cm3) | Tumor weight (g) | Inhibitory ratio (%, w/w) |
|---|---|---|---|---|
| a Data shown are the mean ± SD of tumor weights or volumes for each group of mice (n = 8).b P < 0.01 vs. control. | ||||
| Control | 0.4 mL per mouse | 1.274 ± 0.205 | 1.04 ± 0.31 | |
| 6 | 7.5 mg kg−1 | 0.645 ± 0.147b | 0.52 ± 0.12b | 50.18 |
| 15 mg kg−1 | 0.589 ± 0.140b | 0.41 ± 0.10b | 60.98 | |
3-Nitrotyrosine formation induced by 6 in vitro and in vivo
The generation of highly reactive nitrogen species (RNS) such as peroxynitrite (ONOO−) has been regard as one of the main mechanisms underlying the cytotoxicity of NO against cancer cells.24 ONOO− is capable of nitrating tyrosine residues in many proteins, forming 3-nitrotyrosine (3-NT),25 which is an indirect marker for ONOO− production, as well as an indicator for NO-mediated cell damage.26 Mitochondrial proteins such as cytochrome C (Cyt-c) are important targets for nitration.27 Meanwhile, the nitration of tyrosine residues in Cyt-c can lead to alteration of protein function and cell apoptosis. Accordingly, 3-NT levels of mitochondrial Cyt-c in 6-treated HCT-116 cells were measured using Western blot assay. As shown in Fig. 6a, 6 at 4 μM significantly increased the 3-NT levels in HCT-116 cells, similarly to JS-K and PABA/NO. As expected, pretreatment of cancer cells with either NO scavenger carboxy-PTIO (25 μM, 1 h) or GSTπ inhibitor EA (30 μM, 1 h) led to diminished 3-NT formation by 6. These results demonstrated the ability of 6 to release NO into the mitochondria of colon cancer cells under the catalysis of GSTπ. It was further found that 6 induced 3-NT formation in a dose-dependent manner (Fig. 6b). Moreover, we also observed that treatment with 6 at 15 mg kg−1 for three weeks doubled the amounts of 3-NT in a xenograft mouse model (Fig. 6c).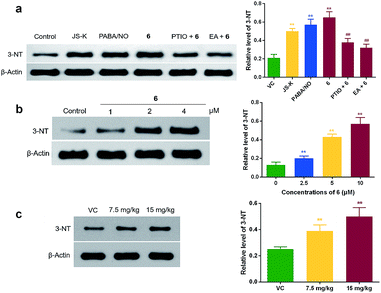 | ||
| Fig. 6 Compound 6 induced the formation of 3-NT in mitochondrial Cyt-c. (a) HCT-116 cells were treated with 4 μM of JS-K, PABA/NO, or 6 for 24 h. Pretreatment of HCT-116 cells with carboxy-PTIO (25 μM, 1 h) or EA (30 μM, 1 h) were used to validate the roles of NO and GSTπ in 3-NT formation. The levels of 3-NT produced in mitochondrial Cyt-c were detected by Western blot assay using specific 3-NT antibody. (b) HCT-116 cells were treated with indicated concentration of 6 for 24 h, the 3-NT levels were detected by Western blot assay. (c) In vivo 3-NT levels were measured in xenografts harvested from above described mice study (Fig. 5). Data are expressed as means + SD from three separate experiments. **P < 0.01 vs. control, ##P < 0.01 vs. 6 alone. | ||
Effects of 6 on Wnt/β-catenin pathway
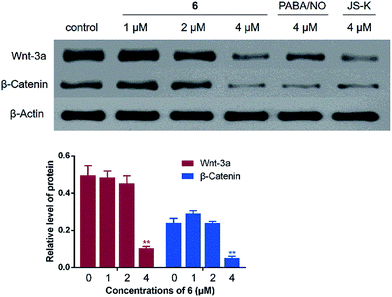
Wnt/β-catenin signaling plays an essential role in the tumor genesis.28 In normal cells, β-catenin degradation is under the control of Wnt signaling. However, mutation in β-catenin is often observed in colon cancer, which stabilizes β-catenin and constitutively activates Wnt signaling, leading to hyperactive proliferation of cancer cells.29 Thus, Western blot assay was then performed to evaluate the effects of 6 on β-catenin and Wnt-3a, one member of the Wnt protein family. It was found that the relative expression levels of Wnt-3a and β-catenin in HCT-116 cells were remarkably decreased by treatment with 4 μM of 6, slightly superior to the effects of JS-K and PABA/NO at the same concentration (Fig. 7).Effects of 6 on COX-2/PGE2 pathway
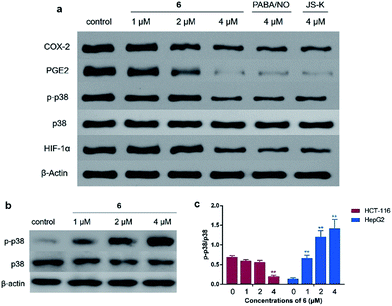
The COX/PG axis represents another important pathway involved in colon cancer pathogenesis. There is evidence showing that COX-2 and PGE2 expression is up-regulated in colon carcinomas, resulting in increased tumor cell invasiveness and aggressiveness. Pharmacological inhibitors of the COX-2/PGE2 axis reduce the occurrence of colon cancer in both experimental models and humans.30–32 Western blot assay was conducted to examine the effects of 6 on COX-2/PGE2 pathway. The results showed that incubation of HCT-116 cells with 6 led to a concentration-dependent reduction of both COX-2 and PGE2 protein levels (Fig. 8a).The phosphorylation of mitogen-activated protein kinase p38 was reported to be responsible for the COX-2 expression in cancer cells, and p38-MAPK pathway inhibitors are able to suppress the COX-2 expression in colon cancer cells.33–35 In consistence with these statements, it was observed that 6 at 4 μM significantly inhibited p38 phosphorylation (Fig. 8a). This result is interesting and thought-provoking, because NO stimuli, generally speaking, is supposed to activate the MAPK pathway by increase the phosphorylation of MAPK family members such as ERK, JNK, and p38. This MAPK-activating effect was previously observed with both PABA/NO13 and our NO–OA hybrids.16,17 Therefore, the current results suggest that NO might exert a unique effect on p38 activation in colon cancer cells compared with that in other cells. To test this hypothesis, HepG2 cells were treated with various concentrations of 6, and the phosphorylated p38 (p-p38) levels were detected using Western blot. As opposed to the case in HCT-116 cells, treatment of HepG2 cells with 6 induced p38 phosphorylation (Fig. 8b and c).
Moreover, it is known that COX-2 up-regulation in colon cancer was associated with hypoxia-inducible factor-1α (HIF-1α) induction.36 In addition, increased levels of PGE2 in hypoxic colorectal tumor cells enhance HIF-1 transcriptional activity by activating MAPK pathway.36 Accordingly, Western blot assay was performed to examine the effects of 6 on COX-2 centered signaling loop in colon cancer cells. The results indicated that HIF-1α expression in HCT-116 cells was suppressed by 4 μM of 6 (Fig. 8a). Taken together, 6 negatively regulated the COX-2/PGE2 pathway and related signaling loop in colon cancer cells.
Conclusion
The potential use of NO-releasing agents in anticancer field has drawn dramatic attention since first reported in 1987.37 Notably, several cancer cell lines were proved to be especially sensitive to NO-induced cytotoxic effects.38 Furthermore, by utilizing cancer-specific features, a much wider range of cancer cells could be efficiently targeted by various NO donors such as dizeniumdiolates.39 We synthesized and biologically evaluated a new group of dizeniumdiolate-based NO-releasing derivatives of OA. MTT assay revealed that 6 was the most potent in inhibiting colon cancer cell proliferation, while sparing normal colon cells. This was demonstrated to be mediated by the selective release of high levels of NO in colon cancer cells but not in normal cells. Importantly, 6 was resistant to GSH-induced decomposition in the absence of GSTπ, and was preferably catalyzed by GSTπ to produce NO rather than by GSTα. The anticancer activity of 6 was further confirmed from its tumor-curtailing effects in a mouse xenograft model.In addition to the downstream effects such as the formation of 3-NT in vitroand in vivo, NO generated by 6 was able to regulate several colon cancer cell-related signaling pathways. For example, the expression levels of Wnt-3a and β-catenin in HCT-116 cells were reduced by treatment with 6. Moreover, the over-expression of COX-2 and PGE2, well known hallmarks in colon cancer, was also negatively regulated by 6. This was accompanied by inhibition of upstream signalling of COX-2/PGE2 axis including HIF-1α and p38 MAPK phosphorylation. Interestingly, the suppressive effect of 6 on p38 activation was only observed in colon cancer cells instead of HCC cells, suggesting its unique mechanism of action in colonic carcinoma.
In summary, 6 as an NO-releasing derivative of OA can be selectively activated by GSTπ over-expressed in colon cancer cells, and effectively regulate colon cancer-related proteins upon activation. As shown in our study, multiple mechanisms might be associated with the inhibitory activity of 6 against colon cancer cells, and several colonic hallmarks are closely involved. Firstly, NO generates ONOO−, which then nitrates tyrosine residues of Cyt-c in mitochondria to form 3-NT. Secondly, 6 negatively regulates the Wnt/β-catenin pathway in colon cancer cells. Thirdly, 6 inhibits the COX-2 centered signalling loop in colon cancer cells. All in all, our findings suggest that 6 might be a potential anti-colon cancer agent, deserving further investigation.
Experimental
General
Melting points were determined on a Mel-TEMP II melting point apparatus and were uncorrected. 1H NMR and 13C NMR spectra were recorded with a Bruker Avance 300 MHz spectrometer at 300 K, using TMS as an internal standard. MS spectra were recorded on a Shimadzu GC-MS 2010 (EI) or a Mariner Mass Spectrum (ESI) instrument. Analytical and preparative TLC was performed on silica gel (200–300 mesh) GF/UV 254 plates, and the chromatograms were visualized under UV light at 254 and 365 nm. All solvents were reagent grade and, when necessary, were purified and dried by standard methods. The purity of all compounds tested was characterized by HPLC analysis (LC-10A HPLC system consisting of LC-10ATvp pumps and an SPD-10Avp UV detector) and high resolution mass spectrometry (Agilent Technologies LC/MSD TOF). Individual compounds with a purity of >95% were used for subsequent experiments (see the ESI†).Sodium diazeniumdiolates 2a–f were prepared according to the method described previously.21 Compound 1, 4, and 14 were synthesized using the reported protocols.16,22 The preparative procedures and spectral data for compounds 5–9, 15a, 15b, 16a and 16b are provided in the ESI.†
General procedure for the preparation of compounds 5–9
To a DMF (8 mL) solution of 14 (745 mg, 1 mmol) and 3b–f (1 mmol) was added Et3N (0.14 mL, 1 mmol), and the resulting solution was stirred at room temperature for 2 h. The mixture was then poured into water (100 mL) prior to extraction with ethyl acetate (3 × 60 mL). The organic extracts were combined and washed sequentially with 1 N HCl (3 × 100 mL) and a saturated NaCl solution, and the organic fraction was dried over sodium sulfate. After removal of the solvent, the crude product was purified by chromatography [1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 (v/v) MeOH–CH2Cl2] to give 5–9.
15 (v/v) MeOH–CH2Cl2] to give 5–9.
Compound 5
The title compound was obtained in 74% yield as a yellow solid: mp 152–154 °C; 1H NMR (d6-DMSO, 300 MHz) δ 0.68 (s, 3H, CH3), 0.78 (s, 6H, 2 × CH3), 0.88 (s, 9H, 3 × CH3), 1.09 (s, 3H, CH3), 2.07–2.27 (m, 4H, prolinol), 2.61–2.81 (m, 5H, piperazine, C18-H), 3.34 (brs, 4H, piperazine), 3.58–3.78 (m, 5H), 3.90–4.11 (m, 5H, prolinol), 4.57–4.69 (m, 2H, OH, 3α-H), 4.76 (d, J = 5.3 Hz, 1H, OH), 4.96 (d, J = 5.0 Hz, 1H, OH), 5.16 (brs, 1H, C12-H), 5.21 (d, J = 7.5 Hz, 1H), 7.20 (s, 1H, ArH), 8.65 (s, 1H, ArH); 13C NMR (d6-DMSO, 75 MHz) δ 175.22, 170.73, 154.65, 148.12, 143.43, 127.26, 125.70, 125.51, 121.47, 98.75, 94.56, 80.54, 75.85, 73.35, 69.41, 67.68, 65.16, 64.98, 59.85, 58.07, 54.47, 51.46, 50.78, 46.84, 45.85, 45.50, 41.22, 41.17, 37.51, 37.20, 36.38, 33.18, 32.68, 32.05, 32.04, 31.56, 30.23, 28.97, 27.60, 27.12, 25.46, 23.30, 23.06, 22.86, 22.45, 17.69, 16.58, 16.49, 15.00; ESI-MS 1070.6 [M + H]+, 1092.4 [M + Na]+; HRMS calculated for C53H80N7O16 [M + H]+ 1070.5662, found 1070.5666 (parts per million error of 0.4).Compound 6
The title compound was obtained in 69% yield as a yellow solid: mp 141–143 °C; 1H NMR (d6-DMSO, 300 MHz) δ 0.69 (s, 3H, CH3), 0.84 (s, 6H, 2 × CH3), 0.88 (s, 9H, 3 × CH3), 1.11 (s, 3H, CH3), 1.97–2.01 (m, 2H, piperidine), 2.60–2.81 (m, 5H, piperazine, C18-H), 3.01 (t, J = 9.8 Hz, 2H, piperidine), 3.18 (brs, 4H, piperazine), 3.40–3.48 (m, 4H), 3.49–3.57 (m, 1H), 3.69–3.74 (m, 2H, piperidine), 4.40–4.47 (m, 1H, 3α-H), 4.49 (d, J = 4.2 Hz, 1H, OH), 4.54–4.56 (m, 1H, piperidine), 4.75 (d, J = 5.1 Hz, 1H, OH), 4.79 (d, J = 4.2 Hz, 1H, OH), 4.96 (d, J = 5.7 Hz, 1H, OH), 5.17 (brs, 1H, C12-H), 5.22 (d, J = 7.8 Hz, 1H), 6.42 (s, 1H, ArH), 8.50 (s, 1H, ArH); 13C NMR (d6-DMSO, 75 MHz) δ 176.82, 169.58, 161.54, 157.98, 143.49, 126.64, 122.24, 110.76, 110.52, 94.80, 75.80, 73.74, 69.83, 68.74, 60.73, 55.31, 51.65, 50.94, 49.65, 45.81, 41.53, 41.18, 39.30, 37.95, 37.43, 36.69, 35.73, 33.48, 32.43, 32.13, 31.75, 30.42, 30.13, 27.48, 27.34, 24.99, 23.15, 22.61, 17.95, 16.36, 15.91, 14.60; ESI-MS 1092.6 [M + Na]+, 1108.6 [M + K]+; HRMS calculated for C53H80N7O16 [M + H]+ 1070.5662, found 1070.5665 (parts per million error of 0.3).Compound 7
The title compound was obtained in 71% yield as a yellow solid: mp 152–155 °C; 1H NMR (d6-DMSO, 300 MHz) δ 0.65 (s, 3H, CH3), 0.72 (s, 6H, 2 × CH3), 0.83 (s, 9H, 3 × CH3), 1.04 (s, 3H, CH3), 2.65 (t, J = 5.2 Hz, 2H, NCH2CH2OH), 2.68–2.75 (m, 5H, piperazine, C18-H), 2.82–2.85 (m, 4H, piperazine), 3.54 (brs, 4H, piperazine), 3.67–3.79 (m, 11H), 4.44–4.48 (m, 3H, 2 × OH, 3α-H), 4.70 (d, J = 4.2 Hz, 1H, OH), 4.91 (d, J = 5.6 Hz, 1H, OH), 5.12 (brs, 1H, C12-H), 5.18 (d, J = 7.5 Hz, 1H), 5.76 (s, 1H, OH), 6.89 (s, 1H, ArH), 8.79 (s, 1H, ArH); 13C NMR (d6-DMSO, 75 MHz) δ 175.26, 170.91, 154.39, 148.15, 143.48, 127.33, 125.95, 125.63, 121.49, 99.40, 94.58, 80.59, 75.89, 73.36, 69.41, 67.71, 59.95, 59.87, 58.07, 57.62, 54.53, 52.86, 49.85, 49.79, 46.88, 45.87, 45.51, 44.95, 41.24, 40.69, 37.56, 37.23, 36.42, 33.09, 32.71, 32.06, 31.59, 30.27, 27.64, 27.14, 25.48, 23.33, 23.10, 22.88, 22.46, 17.71, 16.61, 16.54, 15.03, 14.98; ESI-MS 1099.4 [M + H]+, 1121.4 [M + Na]+; HRMS calculated for C54H83N8O16 [M + H]+ 1099.5849, found 1099.5846 (parts per million error of 0.3).Compound 8
The title compound was obtained in 75% yield as a yellow solid: mp 137–140 °C; 1H NMR (d6-DMSO, 300 MHz) δ 0.69 (s, 3H, CH3), 0.82 (s, 6H, 2 × CH3), 0.88 (s, 9H, 3 × CH3), 1.10 (s, 3H, CH3), 1.91–1.97 (m, 4H, pyrrolidine), 2.70–2.77 (m, 5H, piperazine, C18-H), 3.26 (brs, 4H, piperazine), 3.33–3.51 (m, 4H, pyrrolidine), 3.70 (brs, 5H), 4.45–4.56 (m, 3H, 2 × OH, 3α-H), 4.51 (d, J = 4.3 Hz, OH), 4.77 (d, J = 5.3 Hz, 1H, OH), 4.98 (d, J = 5.4 Hz, 1H, OH), 5.17 (brs, 1H, C12-H), 5.21 (d, J = 7.6 Hz, 1H), 7.10 (s, 1H, ArH), 8.64 (s, 1H, ArH); 13C NMR (d6-DMSO, 75 MHz) δ 175.27, 170.77, 154.94, 148.19, 143.47, 127.35, 125.65, 125.56, 121.51, 98.48, 94.59, 80.57, 75.89, 73.37, 69.43, 67.72, 59.70, 57.62, 54.48, 51.41, 46.85, 45.89, 45.53, 41.26, 37.54, 37.24, 36.42, 33.02, 32.71, 32.08, 31.60, 30.27, 27.64, 27.16, 24.03, 17.72, 16.62, 16.53, 15.04, 14.05; ESI-MS 1040.4 [M + H]+, 1162.4 [M + Na]+; HRMS calculated for C52H78N7O15 [M + H]+ 1040.5478, found 1040.5481 (parts per million error of 0.3).Compound 9
The title compound was obtained in 70% yield as a yellow solid: mp 139–142 °C; 1H NMR (d6-DMSO, 300 MHz) δ 0.68 (s, 3H, CH3), 0.73 (s, 3H, CH3), 0.80 (s, 3H, CH3), 0.87 (s, 9H, 3 × CH3), 1.09 (s, 3H, CH3), 1.90–1.99 (m, 6H, piperidine), 2.70–2.77 (m, 5H, piperazine, C18-H), 3.24 (brs, 4H, piperazine), 3.37–3.52 (m, 5H), 3.72 (t, J = 5.6 Hz, 4H, piperidine), 4.35–4.46 (m, 3H, 2 × OH, 3α-H), 5.16 (brs, 1H, C12-H), 5.21 (d, J = 7.8 Hz, 1H), 6.77 (s, 1H, ArH), 8.90 (s, 1H, ArH); 13C NMR (d6-DMSO, 75 MHz) δ 175.26, 168.50, 154.74, 148.35, 143.47, 127.16, 125.95, 125.75, 121.48, 99.16, 94.58, 81.69, 75.88, 73.38, 69.43, 67.71, 57.62, 54.82, 54.69, 46.81, 46.32, 45.88, 45.53, 41.24, 40.70, 39.61, 38.34, 37.45, 37.28, 36.53, 36.39, 32.70, 32.04, 31.60, 30.26, 27.62, 27.15, 25.50, 24.33, 23.95, 23.32, 23.10, 22.88, 22.47, 17.67, 16.60, 16.36, 15.03; ESI-MS 1054.4 [M + H]+, 1176.4 [M + Na]+; HRMS calculated for C53H80N7O15 [M + H]+ 1054.5634, found 1040.5636 (parts per million error of 0.2).General procedure for the preparation of compounds 10–13
Compounds 15a and 15b were individually dissolved in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of CH2Cl2 and MeOH. A catalytic amount of 10% Pd–C was added to the solution, and the mixture was stirred under hydrogen at room temperature for 10 h. After filtration through Celite, the filtrate was concentrated to afford the corresponding primary amine that was used directly without further purification. A mixture of the primary amine (1 mmol), 3a or 3c (1 mmol), and Na2CO3 (1 mmol) in anhydrous THF was stirred at room temperature until the starting material was totally consumed as indicated by TLC. The remaining solid was filtered off, the filtrate evaporated in vacuo, and the resulting residue purified by column chromatography [1
1 mixture of CH2Cl2 and MeOH. A catalytic amount of 10% Pd–C was added to the solution, and the mixture was stirred under hydrogen at room temperature for 10 h. After filtration through Celite, the filtrate was concentrated to afford the corresponding primary amine that was used directly without further purification. A mixture of the primary amine (1 mmol), 3a or 3c (1 mmol), and Na2CO3 (1 mmol) in anhydrous THF was stirred at room temperature until the starting material was totally consumed as indicated by TLC. The remaining solid was filtered off, the filtrate evaporated in vacuo, and the resulting residue purified by column chromatography [1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (v/v) MeOH–CH2Cl2] to afford the respective title product.
20 (v/v) MeOH–CH2Cl2] to afford the respective title product.
Compound 10
The title compound was obtained in 82% yield as a yellow solid: mp 127–131 °C; 1H NMR (d6-DMSO, 300 MHz) δ 0.68 (s, 6H, 2 × CH3), 0.75 (s, 3H, CH3), 0.83 (s, 3H, CH3), 0.88 (s, 6H, 2 × CH3), 1.09 (s, 3H, CH3), 1.52–1.82 (m, 4H, proline), 2.73–2.76 (m, 1H, C18-H), 3.31 (s, 3H, NCH3), 3.37–3.58 (m, 7H), 3.64–3.67 (m, 2H, NCH2CH2OH), 3.71–3.72 (m, 1H), 3.73–3.74 (m, 2H, NCH2CH2OH), 4.30 (m, 1H, proline),4.35–4.50 (m, 3H, 2 × OH, 3α-H), 5.16 (brs, 1H, C12-H), 5.21 (d, J = 7.8 Hz, 1H), 6.77 (s, 1H, ArH), 8.90 (s, 1H, ArH); ESI-MS 1015.5 [M + H]+; 13C NMR (d6-DMSO, 75 MHz) δ 175.24, 172.58, 154.91, 148.17, 143.45, 127.35, 125.63, 125.54, 121.47, 98.49, 94.56, 80.54, 75.86, 73.34, 69.41, 67.68, 61.20, 59.85, 57.58, 54.71, 54.45, 51.87, 46.82, 45.86, 45.48, 41.23, 40.68, 37.50, 37.21, 36.39, 33.18, 33.00, 32.68, 32.03, 31.58, 30.25, 28.95, 27.61, 27.12, 25.47, 25.01, 23.31, 23.12, 23.05, 22.85, 22.46, 22.43, 17.68, 16.59, 16.50, 15.02; HRMS calculated for C50H75N6O16 [M + H]+ 1015.5161, found 1015.5163 (parts per million error of 0.2).Compound 11
The title compound was obtained in 75% yield as a yellow solid: mp 130–132 °C; 1H NMR (d6-DMSO, 300 MHz) δ 0.68 (s, 3H, CH3), 0.79 (s, 3H, CH3), 0.85 (s, 3H, CH3), 0.88 (s, 9H, 3 × CH3), 1.09 (s, 3H, CH3), 2.73–2.77 (m, 1H, C18-H), 2.96 (s, 3H, NCH3), 3.33 (s, 3H, NCH3), 3.38–3.52 (m, 5H), 3.65–3.68 (m, 2H, NCH2CH2OH), 3.70–3.72 (m, 1H), 3.73–3.74 (m, 2H, NCH2CH2OH), 4.37–4.53 (m, 5H, 2 × OH, 3α-H, NHCH2COO), 5.16 (brs, 1H, C12-H), 5.20 (d, J = 7.7 Hz, 1H), 6.93 (s, 1H, ArH), 8.63 (s, 1H, ArH); ESI-MS 1011.5 [M + Na]+; 13C NMR (d6-DMSO, 75 MHz) δ 175.25, 172.19, 154.85, 148.41, 143.45, 127.41, 125.69, 125.36, 121.48, 98.43, 94.56, 80.10, 75.87, 73.34, 69.40, 67.69, 59.86, 57.60, 54.75, 54.52, 46.84, 45.86, 45.50, 42.11, 41.23, 40.69, 37.54, 37.23, 36.40, 33.17, 32.69, 32.06, 31.57, 30.92, 30.25, 27.69, 27.12, 25.46, 23.31, 23.09, 23.04, 22.87, 22.45, 17.71, 16.60, 16.57, 15.02; HRMS calculated for C48H73N6O16 [M + H]+ 989.5005, found 989.5008 (parts per million error of 0.3).Compound 12
The title compound was obtained in 65% yield as a yellow solid: mp 142–143 °C; 1H NMR (d6-DMSO, 300 MHz) δ 0.67 (s, 3H, CH3), 0.79 (s, 6H, 2 × CH3), 0.87 (s, 9H, 3 × CH3), 1.09 (s, 3H, CH3), 2.72–2.77 (m, 1H, C18-H), 3.40–3.67 (m, 8H), 3.73–3.82 (m, 4H, piperidine), 4.37–4.40 (m, 1H, proline), 4.49–4.51 (m, 1H, 3α-H), 4.86 (d, J = 3.7 Hz, 1H, OH), 4.96 (d, J = 5.6 Hz, 1H, OH), 5.16 (brs, 1H, C12-H), 5.20 (d, J = 7.7 Hz, 1H), 6.96 (s, 1H, ArH), 8.63 (s, 1H, ArH); 13C NMR (d6-DMSO, 75 MHz) δ 175.23, 172.60, 154.63, 148.10, 143.44, 127.29, 125.76, 125.66, 121.47, 98.83, 94.58, 80.55, 75.87, 73.39, 69.43, 67.70, 67.02, 60.23, 59.87, 54.89, 54.51, 51.84, 46.88, 46.19, 46.06, 45.86, 45.52, 41.22, 37.54, 37.21, 36.40, 32.97, 32.69, 32.07, 31.59, 30.24, 29.12, 27.62, 27.13, 25.00, 25.46, 23.31, 23.07, 22.87, 22.45, 17.70, 16.59, 16.49, 15.01; ESI-MS 1063.5 [M + Na]+; HRMS calculated for C52H77N6O16 [M + H]+ 1041.5318, found 1041.5322 (parts per million error of 0.4).Compound 13
The title compound was obtained in 72% yield as a yellow solid: mp 152–155 °C; 1H NMR (d6-DMSO, 300 MHz) δ 0.68 (s, 3H, CH3), 0.77 (s, 3H, CH3), 0.83 (s, 3H, CH3), 0.88 (s, 9H, 3 × CH3), 1.09 (s, 3H, CH3), 2.73–2.77 (m, 1H, C18-H), 2.94 (s, 3H, NCH3), 3.39–3.51 (m, 5H), 3.63–3.73 (m, 2H, piperidine), 3.74–3.86 (m, 2H, piperidine), 4.39–4.55 (m, 3H, piperidine, 3α-H, OH), 4.78 (d, J = 3.7 Hz, 1H, OH), 4.98 (d, J = 5.6 Hz, 1H, OH), 5.17 (brs, 1H, C12-H), 5.22 (d, J = 7.8 Hz, 1H), 6.95 (s, 1H, ArH), 8.62 (s, 1H, ArH); 13C NMR (d6-DMSO, 75 MHz) δ 175.25, 170.64, 154.89, 147.58, 143.46, 127.26, 126.22, 125.93, 121.45, 99.15, 94.56, 82.02, 75.86, 73.34, 69.40, 67.68, 67.05, 54.90, 54.76, 54.71, 54.33, 46.77, 45.86, 45.50, 41.23, 40.68, 37.37, 37.19, 36.35, 33.89, 32.02, 32.00, 31.57, 30.25, 27.54, 27.13, 25.48, 24.69, 23.31, 23.05, 22.87, 22.46, 21.99, 17.62, 16.58, 16.35, 14.97; ESI-MS 1037.5 [M + Na]+; HRMS calculated for C50H75N6O16 [M + H]+ 1015.5161, found 1015.5165 (parts per million error of 0.4).MTT assay
The inhibitory effects on cell proliferation of test compounds were investigated by the MTT method. HT-29, HCT-15, HCT-116, or normal human colon cell line CCD-841 cells at a final density of 1.0 × 104 cells per well were placed in 96-well cell plates overnight and treated with or without different concentrations of test compounds for various periods of time. During the last 4 h culture, the cells were exposed to MTT (5 mg mL−1), and the resulting formazan crystals were dissolved in 150 μL of DMSO and measured using a spectrophotometer (Tecan) at a test wavelength of 570 nm. Experiments were conducted in triplicate. Inhibition rate (%) = [(Acontrol − Atreated)/Acontrol] × 100%.Nitrite measurement by the Griess assay
The levels of NO generated by individual compounds in the cells are presented as nitrite and were determined by the colorimetric assay using the nitrite colorimetric assay kit (Beyotime, Nanjing, China), according to the manufacturer's instructions. Briefly, HCT-116 or CCD-841 cells were treated with the indicated concentrations of test compounds, and the nitrite contents of the cell lysates were detected by the Griess assay. The absorbance was read at 540 nm on a spectrophotometer (Smart spec, Bio-Rad). The amount of nitrite in the lysates was calculated using a NaNO2 standard curve in accordance with the manufacturer's instructions (Beyotime).Stability in absence of GSTπ monitored by HPLC
Individual test compounds (40 μM) were incubated with a 0.1 M PBS (pH 7.4) buffer containing 4 mM of GSH, BSA, or homocystein. The mixture was allowed to react at 37 °C, and the reaction was monitored by HPLC (Shimadzu) at the indicated time points. The HPLC conditions utilized consisted of a 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 acetonitrile–water mixture with 0.1% formic acid and a flow rate of 1 mL min−1. Detection wavelengths were 339, 298, and 310 nm for 6, JS-K, and PABA/NO, respectively.
30 acetonitrile–water mixture with 0.1% formic acid and a flow rate of 1 mL min−1. Detection wavelengths were 339, 298, and 310 nm for 6, JS-K, and PABA/NO, respectively.
Enzymatic decomposition of 6 in the presence of GST
To a 200 μL 0.1 M PBS (pH 7.4) reaction mixture containing 1 mM GSH and 1 μg of GSTπ (PROSPEC) or GSTα (PROSPEC) was added an appropriate amount of 21 (DMSO stock solution). The final concentrations of 6 varied from 0 to 60 μM, and its decomposition behavior was monitored at room temperature using a microplate reader (SH-1000, Shimadzu) at its λmax (339 nm). The parameters were calculated using the Michaelis–Menten equation.In vivo antitumor effects of 6
Male BALB/c nude mice 3–5 weeks of age (Silaike Experiment Animal Co.,Ltd, Shanghai, China) were inoculated subcutaneously with 1 × 107 SMMC-7721 cells. After the formation of a solid tumor with a volume of about 75 mm3, the tumor-bearing mice were randomly divided into three groups with eight mice in each group. The groups with compound 6 treatment received two dosages (7.5 or 15 mg kg−1) by intravenous injection every day. At the end of the experiment, the mice were sacrificed, their tumors were dissected, and the tumor size and weight were measured. The tumor inhibitory ratio was calculated by the following formula: tumor inhibitory ratio (%) = [(Wcontrol − Wtreated)/Wcontrol] × 100%. Wtreated and Wcontrol were the average tumor weights of the treated and control mice, respectively. The tumor diameters were measured with calipers, and the tumor volume was calculated by the formula| V (mm3) = d2 × D/2, |
Western blot assay
HCT-116 cells were extracted with a cell lysis buffer (Beyotime). The total protein concentration was measured and adjusted to equal concentrations across different samples. The protein was separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore). The PVDF membranes were incubated with the indicated primary antibodies overnight at 4 °C and then incubated with the secondary antibodies conjugated to horseradish peroxidase. The proteins were visualized by a Keygen ECL system (Kaiji, Nanjing, China) and scanned with a Clinx ChemiScope chemiluminescence imaging system (Gel Catcher 2850). The relative optical densities of the specific proteins were determined with a ChemiScope analysis program.Acknowledgements
This study was supported by grants from National Natural Science Foundation of China (81273378 and 81202408). The GSTπ inhibitor etacrynic acid (EA) was a generous gift from Prof. Guisen Zhao (Shandong University, Jinan, China).Notes and references
- C. N. Arnold, A. Goel, H. E. Blum and C. R. Boland, Cancer, 2005, 104, 2035–2047 CrossRef CAS PubMed.
- D. Colussi, G. Brandi, F. Bazzoli and L. Ricciardiello, Int. J. Mol. Sci., 2013, 14, 16365–16385 CrossRef PubMed.
- B. Bonavida, S. Baritaki, S. Huerta-Yepez, M. I. Vega, D. Chatterjee and K. Yeung, Nitric Oxide, 2008, 19, 152–157 CrossRef CAS PubMed.
- D. Fukumura, S. Kashiwagi and R. K. Jain, Nat. Rev. Cancer, 2006, 6, 521–534 CrossRef CAS PubMed.
- S. Huerta, S. Chilka and B. Bonavida, Int. J. Oncol., 2008, 33, 909–927 CAS.
- J. L. Williams, P. Ji, N. Ouyang, L. Kopelovich and B. Rigas, Exp. Cell Res., 2011, 317, 1359–1367 CrossRef CAS PubMed.
- M. Mojic, S. Mijatovic, D. Maksimovic-Ivanic, D. Miljkovic, S. Stosic-Grujicic, M. Stankovic, K. Mangano, S. Travali, M. Donia, P. Fagone, M. B. Zocca, Y. Al-Abed, J. A. McCubrey and F. Nicoletti, Mol. Pharmacol., 2012, 82, 700–710 CrossRef CAS PubMed.
- G. Zhao and X. Wang, Curr. Med. Chem., 2006, 13, 1461–1471 CrossRef CAS.
- C. C. Chao, Y. T. Huang, C. M. Ma, W. Y. Chou and S. Lin-Chao, Mol. Pharmacol., 1992, 41, 69–75 CAS.
- M. L. Clapper, S. J. Hoffman and K. D. Tew, Biochim. Biophys. Acta, 1991, 1096, 209–216 CrossRef CAS.
- A. D. Lewis, L. M. Forrester, J. D. Hayes, C. J. Wareing, J. Carmichael, A. L. Harris, M. Mooghen and C. R. Wolf, Br. J. Cancer, 1989, 60, 327–331 CrossRef CAS.
- K. A. Naidu, A. Nasir, H. Pinkas, H. E. Kaiser, P. Brady and D. Coppola, In Vivo, 2003, 17, 479–482 CAS.
- V. J. Findlay, D. M. Townsend, J. E. Saavedra, G. S. Buzard, M. L. Citro, L. K. Keefer, X. Ji and K. D. Tew, Mol. Pharmacol., 2004, 65, 1070–1079 CrossRef CAS PubMed.
- A. E. Maciag, J. E. Saavedra and H. Chakrapani, Anti-Cancer Agents Med. Chem., 2009, 9, 798–803 CrossRef CAS.
- P. J. Shami, J. E. Saavedra, C. L. Bonifant, J. Chu, V. Udupi, S. Malaviya, B. I. Carr, S. Kar, M. Wang, L. Jia, X. Ji and L. K. Keefer, J. Med. Chem., 2006, 49, 4356–4366 CrossRef CAS PubMed.
- J. Fu, L. Liu, Z. Huang, Y. Lai, H. Ji, S. Peng, J. Tian and Y. Zhang, J. Med. Chem., 2013, 56, 4641–4655 CrossRef CAS PubMed.
- L. Liu, J. Fu, T. Li, R. Cui, J. Ling, X. Yu, H. Ji and Y. Zhang, Eur. J. Pharmacol., 2012, 691, 61–68 CrossRef CAS PubMed.
- N. B. Janakiram, C. Indranie, S. V. Malisetty, P. Jagan, V. E. Steele and C. V. Rao, Pharm. Res., 2008, 25, 2151–2157 CrossRef CAS PubMed.
- S. M. Kim, I. H. Jeong, M. S. Yim, M. K. Chae, H. N. Kim, D. K. Kim, C. M. Kang, Y. S. Choe, C. Lee and E. K. Ryu, J. Drug Targeting, 2014, 22, 191–199 CrossRef CAS PubMed.
- J. Li, W. J. Guo and Q. Y. Yang, World J. Gastroenterol., 2002, 8, 493–495 CAS.
- Z. Huang, Y. Zhang, L. Fang, Z. Zhang, Y. Lai, Y. Ding, F. Cao, J. Zhang and S. Peng, Chem. Commun., 2009, 1763–1765 RSC.
- Z. Huang, Y. Zhang, L. Zhao, Y. Jing, Y. Lai, L. Zhang, Q. Guo, S. Yuan, J. Zhang, L. Chen, S. Peng and J. Tian, Org. Biomol. Chem., 2010, 8, 632–639 CAS.
- T. Zhou, A. A. Evans, W. T. London, X. Xia, H. Zou, F. Shen and M. L. Clapper, Cancer Res., 1997, 57, 2749–2753 CAS.
- D. D. Thomas, L. A. Ridnour, J. S. Isenberg, W. Flores-Santana, C. H. Switzer, S. Donzelli, P. Hussain, C. Vecoli, N. Paolocci, S. Ambs, C. A. Colton, C. C. Harris, D. D. Roberts and D. A. Wink, Free Radical Biol. Med., 2008, 45, 18–31 CrossRef CAS PubMed.
- K. A. Hanafy, J. S. Krumenacker and F. Murad, Med. Sci. Monit., 2001, 7, 801–819 CAS.
- C. Herce-Pagliai, S. Kotecha and D. E. Shuker, Nitric Oxide, 1998, 2, 324–336 CrossRef CAS PubMed.
- I. V. Turko, L. Li, K. S. Aulak, D. J. Stuehr, J. Y. Chang and F. Murad, J. Biol. Chem., 2003, 278, 33972–33977 CrossRef CAS PubMed.
- H. Clevers, Cell, 2006, 127, 469–480 CrossRef CAS PubMed.
- P. Polakis, Genes Dev., 2000, 14, 1837–1851 CAS.
- C. H. Koehne and R. N. Dubois, Semin. Oncol., 2004, 31, 12–21 CrossRef CAS PubMed.
- H. Kohno, R. Suzuki, S. Sugie and T. Tanaka, BMC Cancer, 2005, 5, 46 CrossRef PubMed.
- J. B. Tuynman, M. P. Peppelenbosch and D. J. Richel, Crit. Rev. Oncol. Hematol., 2004, 52, 81–101 CrossRef CAS PubMed.
- M. L. Gauthier, C. R. Pickering, C. J. Miller, C. A. Fordyce, K. L. Chew, H. K. Berman and T. D. Tlsty, Cancer Res., 2005, 65, 1792–1799 CrossRef CAS PubMed.
- Z. Guan, L. D. Baier and A. R. Morrison, J. Biol. Chem., 1997, 272, 8083–8089 CrossRef CAS PubMed.
- W. Liu, N. Reinmuth, O. Stoeltzing, A. A. Parikh, C. Tellez, S. Williams, Y. D. Jung, F. Fan, A. Takeda, M. Akagi, M. Bar-Eli, G. E. Gallick and L. M. Ellis, Cancer Res., 2003, 63, 3632–3636 CAS.
- A. Kaidi, D. Qualtrough, A. C. Williams and C. Paraskeva, Cancer Res., 2006, 66, 6683–6691 CrossRef CAS PubMed.
- J. B. Hibbs Jr, R. R. Taintor and Z. Vavrin, Science, 1987, 235, 473–476 CAS.
- P. J. Shami, D. L. Sauls and J. B. Weinberg, Leukemia, 1998, 12, 1461–1466 CAS.
- L. K. Keefer, ACS Chem. Biol., 2011, 6, 1147–1155 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Cytotoxicity of 6 against various cancer cell lines; representative curves showing the decomposition of 6 in pH 7.4 buffer containing 1 mM GSH with or without GST; experimental procedures for 3b–f, 15a, 15b, 16a and 16b; HPLC spectra for the purities of 5–13; NMR spectra of representative compounds. See DOI: 10.1039/c5ra00270b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |