Structure-based virtual screening and ADME/T-based profiling for low molecular weight chemical starting points as p21-activated kinase 4 inhibitors†
Abstract
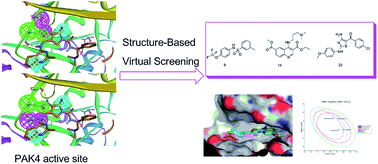
A structure-based virtual screening approach to targeting p21-activated kinase 4 (PAK4) was performed to identify good chemical starting points for medicinal chemistry. A pre-filtrated database was screened against two designed PAK4 pharmacophores, and the pharmacophore search hits were docked into a PAK4 crystal structure. Twenty-seven compounds were then selected for in vitro PAK4 inhibition assay, and results showed three compounds exhibiting a micro-molar IC50 in a dose–response assay. Interactive modes of the three compounds were studied and showed good binding modes in the PAK4 active site. Calculated ADME/T properties of the three hits were also analyzed and showed good drug-like properties. The results of in vitro PAK4 inhibition assay, interactive mode study and ADME/T prediction revealed that the three compounds have potential PAK4 inhibitory activities and can be further optimized and developed as lead compounds.

 Please wait while we load your content...
Please wait while we load your content...