Designed biomolecule–cellulose complexes for palladium recovery and detoxification†
Ian Sofian Yunus and
Shen-Long Tsai*
Department of Chemical Engineering, National Taiwan University of Science and Technology, No. 43 Keelung Rd., Daan Dist., 10607 Taipei, Taiwan. E-mail: stsai@mail.ntust.edu.tw
First published on 12th February 2015
Abstract
An efficient, selective and reusable biosorbent is important for precious metal recovery. This paper examines the recovery of palladium Pd(II) from wastewater on designed biomolecule–cellulose complexes. A genetically engineered fusion protein composed of palladium binding peptides (PdBP) and cellulose binding domains (CBD) was expressed in Escherichia coli and enabled the binding of cellulose for palladium recovery and detoxification. The results of this study indicated that the range of pH levels suitable for PdCBD–cellulose complexes is wide and that the effect of temperature on palladium recovery is insignificant. In addition, the PdCBD–cellulose complexes exhibited good reusability and a high selectivity for Pd(II) recovery. The maximum adsorption capacity of the PdCBD–cellulose was 175.44 mg g−1, indicating a high adsorption capacity for Pd(II). The Langmuir adsorption isotherm was applied to describe the processes for removing Pd(II). The kinetics of the Pd(II) removal were identified as following a pseudo-second order rate equation. Furthermore, the results of a Lemna minor growth inhibition test showed effective detoxification of the designed complexes. The results of this study revealed that the designed biomolecule–cellulose complexes can be used to develop a selective process for recovering and detoxifying precious metals.
Introduction
In 1993, the catalytic converter industry began producing a three-way converter that contains palladium Pd(II) as an alternative to traditional platinum (Pt)–rhodium (Rh) autocatalysts. These noble metals are used to reduce hydrocarbon, carbon monoxide and nitrous oxide in automotive emissions.1 Although large amounts of these gaseous pollutants are removed from emissions by using autocatalysts, their use has led to increased concentrations of platinum group elements (PGEs) in the environment. Although the concentrations of palladium in aquatic systems is usually substantially lower than its acute toxic level, the long-term effects of palladium-bioaccumulation in the aquatic biota can harm humans because humans are the final receptor in the food chain.2 Furthermore, the limited supply and high worldwide demand of palladium have caused extreme price volatility.3 Thus, the development of adsorbents that are capable of efficiently removing and recovering palladium from aqueous solutions or waste solutions is crucial.Conventional physicochemical methods of recovering palladium (such as solvent extraction, membrane filtration, reverse osmosis, chemical precipitation, and ion exchange) have considerable disadvantages, including incomplete removal of metal, lack of specificity, extensive labour and time requirements, and generation of toxic sludge or other waste products that require disposal. Another method is biosorption which is more economical and environmentally friendly than conventional methods.4
Biosorbents such as chitosan,5 lignin,6 tannin,7 and microorganisms8 are effective in palladium ions from aqueous solutions. Although biosorbents have shown high adsorption capacities, developing selective sorbents remains a challenging task. Moreover, microorganism-based biosorbents have many drawbacks such as poor mechanical strength, limited rigidity, and difficulty in separating solids and liquid. Immobilization techniques have been used to overcome the drawbacks of microorganism-based biosorbents.3,9
In our research, we constructed a protein fusion of palladium binding peptides (PdBP) that bind N-terminally to cellulose binding domains (CBD), thereby eliminating the aforementioned problems. PdBP have been reported to size-regulate nanoparticle formation and are known to specifically bind target palladium ions.10 CBDs, which are commonly used in the biotechnology field as an affinity tag protein for immobilizing other proteins in cellulose, binds rapidly, tightly, and specifically to cellulose in wide ranges of pH and temperature.11 Cellulose is a naturally abundant, inexpensive, and chemically inert material with inherently low binding characteristics.12 The unique properties of PdBP and CBD, in addition to the low cost of cellulose, may enable the commercial application of a PdBP–CBD-based adsorbent for recovering palladium ions.
The objectives of this study were (1) to construct a PdBP–CBD-based adsorbent and to study the Pd(II) adsorption behaviour of PdBP–CBD-cellulose as a function of initial pH and temperature; (2) to investigate the effects of the initial concentrations of Pd(II) on Pd(II) adsorption; (3) to delineate the mechanism of Pd(II) adsorption on PdBP–CBD-cellulose; (4) to simulate a Pd(II) adsorption isotherm and to estimate the adsorption capacities; (5) to examine the reusability of PdBP–CBD cellulose; (6) to evaluate the selectivity of PdBP–CBD-cellulose toward metals other than palladium; and (7) to investigate the possibility of using PdBP–CBD-cellulose for detoxifying of palladium polluted water.
Experimental
Plasmid constructions and protein expression
The PdBP–CBD plasmid was constructed in two steps. First, the PdBP was amplified from a pPd-YFP plasmid by using F-XbaI-CBD and R-NdeI-CBD primers (ESI, Table S1†) in PCR. The PdBP gene was then digested using XbaI and NdeI and inserted into a pET-24a (+) vector to form pET24a-PdBP. Second, the CBD gene was amplified from a pCBD-blue plasmid by using F-NdeI-CBD and R-XhoI-CBD primers in PCR. The amplified CBD gene was digested using NdeI and XhoI and then inserted into pET24a-PdBP to form ET24a-PdBP–CBD. A scheme of constructs can be found in ESI, Fig. S1.† Finally, the plasmid was transformed into E. coli BL21 (DE3).Two milliliters of overnight preculture was mixed into 200 mL of Luria–Bertani (LB) medium (10.0 g L−1 tryptone, 5.0 g L−1 yeast extract, 10.0 g L−1 NaCl) supplemented with 50 μg mL−1 kanamycin. When OD600 reached 0.5, the cultures were supplemented with 0.5 mM IPTG and then incubated for 2.5 hours at 30 °C. Cells were harvested through centrifugation at 8500 rpm for 20 min and resuspended in 2 mL of DI water. The cell suspension was lysed through sonication and centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 7 min. The supernatant was heat purified at 75 °C for 30 min and then centrifuged to collect the heat-purified protein. The resulting supernatant was recovered and used for cellulose binding and palladium adsorption studies. The unpurified and purified proteins were subjected to an SDS-PAGE analysis. Protein quantification was performed using the BioRad (Bradford) protein assay kit.
000 rpm for 7 min. The supernatant was heat purified at 75 °C for 30 min and then centrifuged to collect the heat-purified protein. The resulting supernatant was recovered and used for cellulose binding and palladium adsorption studies. The unpurified and purified proteins were subjected to an SDS-PAGE analysis. Protein quantification was performed using the BioRad (Bradford) protein assay kit.
Batch adsorption of palladium
One milliliter of purified PdBP–CBD (8.4 mg mL−1) was then exposed to 0.1 g cellulose which was prewashed with DI water. After 30 min of gentle agitation by using a mechanical mixer at room temperature, the PdBP–CBD-cellulose complexes were recovered through centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 min and repeatedly rinsed with DI water. Subsequently, the adsorption kinetics of Pd(II) on the PdBP–CBD-cellulose was examined by mixing the prepared sorbent with 1.5 mL of 50 mg L−1 Pd(II) solution in 2 mL vials at room temperature. Pd(II) solution was prepared by dissolving Pd(NH3)4Cl2 in DI water. The vials were then shaken using a mechanical mixer. At given time intervals, the mixed solutions were centrifuged at 10
000 rpm for 3 min and repeatedly rinsed with DI water. Subsequently, the adsorption kinetics of Pd(II) on the PdBP–CBD-cellulose was examined by mixing the prepared sorbent with 1.5 mL of 50 mg L−1 Pd(II) solution in 2 mL vials at room temperature. Pd(II) solution was prepared by dissolving Pd(NH3)4Cl2 in DI water. The vials were then shaken using a mechanical mixer. At given time intervals, the mixed solutions were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 min and filtered through a 0.22 μm PVDF filter (Millipore MillexHV) prior to inductively coupled plasma atomic emission spectroscopy (ICP-AES) measurement.
000 rpm for 3 min and filtered through a 0.22 μm PVDF filter (Millipore MillexHV) prior to inductively coupled plasma atomic emission spectroscopy (ICP-AES) measurement.
The ability of PdBP–CBD-cellulose complexes to adsorb palladium in various condition (pH and temperature) was also investigated. HCl and NaOH were used to adjust the pH to the desired value. To determine the selectivity in the adsorption of palladium ions, 10, 50, and 100 mg L−1 of Ca(II), Mg(II), Sr(II), and Pt(IV) were incubated individually with PdBP–CBD-cellulose to replace Pd(II). To investigate the effect of the interfering ions on adsorption, Pd(II) and Pt(IV) were incubated together. After the treatment, the metal ions content in the samples was measured by ICP-AES.
The capacity of the PdBP–CBD-cellulose to adsorb palladium was determined by mixing 0.1 g of the sorbent with 1.5 mL of Pd(II) solutions with concentrations ranging from 5 to 500 mg L−1 in 2 mL vials at room temperature. The pH values of all Pd(II) solutions were adjusted to 3.54 ± 0.2. The adsorption capacity was calculated according to the equation
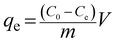 | (1) |
Recovery, reuse and continuous adsorption
PdBP–CBD-cellulose complexes were mixed with 1.5 mL of 10, 50, and 100 mg L−1 Pd(II) solutions in 2 mL vials at room temperature. The used sorbent was washed three times with DI water and the adsorbed Pd(II) ions were recovered using 1.5 mL of a 1 M thiourea solution. After recovery, the PdBP–CBD-cellulose was thoroughly washed with DI water to prepare it for reuse. The reusability of the sorbent was tested for five sorption–desorption cycles.Lemna minor growth inhibition test
A Lemna minor growth inhibition test was performed according to OECD211 guidelines.13 In brief, young and rapidly growing plants without visible chlorosis were collected from field water. Three L. minor growth medium were prepared. First, field-collected water was filtered through a 0.22 μm PVDF filter (Millipore MillexHV) and mixed with Pd(NH3)4Cl2 to a concentration of 10 mg L−1. Second, the filtered field-collected water was mixed with Pd(NH3)4Cl2 to a concentration of 10 mg L−1 and treated with 100 mg PdBP–CBD-cellulose complexes. After adsorption, the supernatant was collected and used as an L. minor growth medium. Third, field-collected water was used as a control. For each sample, 300 μL of the solution was aliquoted into ten 96-well plates. A single frond L. minor was placed into each well. All tests were performed indoor in environmental conditions and three replicates were prepared for each experimental set. The relative plant growth (RG) was monitored for 14 days and calculated using eqn (2):14
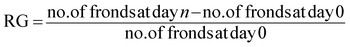 | (2) |
Results and discussion
pH and temperature effects
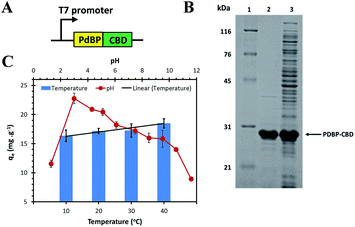
In our system (Fig. 1A), the PdBP was expressed as an in-frame fusion protein with a thermotolerant CBD derived from the clostridium thermocellum cellulosome. The fusion protein can be purified simply by using a heat purification method. The SDS-PAGE result (Fig. 1B) revealed that the size of the fusion protein is approximately 30 kDa. We aimed to develop an adsorbent that binds specifically to Pd(II) and works in a wide-range of adsorption condition.Fig. 1C shows the profile of Pd(II) adsorption on PdBP–CBD-cellulose at different pH and temperature levels. The results indicated that the adsorption capacity of Pd(II) substantially increased when the pH was between 1.3 and 3.08, where it reached the maximum (qe = 22.77 mg g−1) and decreased notably when the pH was between 3.08 and 11.68, indicating that palladium adsorption is favourable at low pH levels. This result was in line with results obtained by other studies on precious metal adsorption, in which higher adsorption capacity for precious metal ions was obtained at low pH.9 In an acidic condition, the Pd(II) adsorption mechanism of PdBP–CBD-cellulose is assumed to be electrostatic attraction and ion exchange.15 To support this assumption, Hydra and Medusa computer program was used to determine the distribution of Pd species under the experimental conditions.16 In the case of hydrochloric solutions, when chloride concentration is higher than 10 mM, PdCl42−, PdCl3−, and PdCl2 represent 85%, 10%, and 5% of total palladium concentration (at 50 ppm), respectively (ESI, Fig. S2 and S3†). For pH lower than 5, anionic and nonionic palladium species represent 95% and 5% of total palladium concentration, respectively. This indicates that in the presence of chloride, palladium ions form chloropalladate anions which interact with protonated amine sties on PdBP–CBD [(RNH3+)2PdCl42−].17 It is found that the binding peptide is positively charged in a wide pH range from 0 to 11 based on the calulation.18 As the pH increase, the distributions of cationic species also increase (ESI, Fig. S2 and S4†). Therefore, there is electrostatic repulsion between the peptide and cationic palladium species, resulting in lower adsorption capacities. This results suggest that the adsorption of cationic species (such as Pd(NH3)42+ and Pd(NH3)32+) are less favourable in high pH. The decrease in adsorption capacity as the pH increases may also be explained by the fewer protonated amine sites available on PdBP–CBD and the fewer chloride anions available.15 However, we observed a substantially low adsorption capacity of Pd(II) at pH 1.3 and 11.68. At pH 1.3, a portion of PdBP–CBD was incapable of binding to the cellulose because the protein denatured, causing some Pd–PdBP–CBD to remain in the supernatant (see ESI, Fig. S5†). The low palladium adsorption capacity at pH 11.68 was attributed to the same reason.
The temperature at which a sorption reaction occurs is seldom of importance, with most reactions being temperature-independent or exothermic reactions.19 In this study, the results showed that the adsorption capacity increased insignificantly as the temperature increased (Fig. 1C), indicating that the adsorption process was endothermic. However, at 20 °C and 30 °C, the temperature did not seem to influence the adsorption performance. In addition, the designed biosorbent was quite stable as no significant decrease in binding efficiency and no detectable protein bleeding could be observed after two months storage at room temperature (25 ± 2 °C). Therefore, all subsequent experiments were conducted at room temperature.
Selectivity and reusability
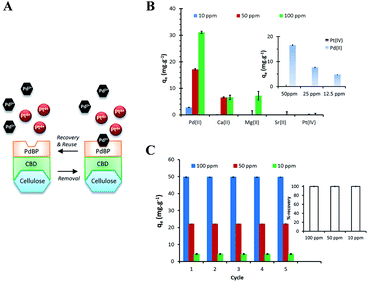
Palladium and platinum have been used in manufacturing automotive catalytic converters for the past few years, causing large amounts of these metals to be deposited in river sediment and lakes. By contrast, Ca(II) and Mg(II) are commonly found in natural water systems.20 Therefore, developing an adsorbent that is selective in adsorbing palladium is crucial. This study investigated the adsorption selectivity of PdBP–CBD-cellulose for Pd(II) ions relative to that for Pt(IV), Ca(II), Mg(II), and Sr(II) ions. As shown in Fig. 2B, the adsorption of Pd(II) was markedly higher than the adsorption of other metal ions. However, because the adsorbent was composed of PdBPs and CBDs, it is not surprising that some non-specific adsorption of other metal ions was observed. Furthermore, in many palladium-polluted waters, palladium is accompanied by platinum. Therefore, the adsorption selectivity of Pd(II) in Pd(II) and Pt(IV) solutions was examined. The inset in Fig. 2B shows that there was no significant adsorption competition between Pd(II) and Pt(IV) ions, indicating a high stability of the designed biosorbent.To examine whether the designer biosorbent can be constantly regenerated and reused, experiments were performed to determine whether the Pd(II) bound to PdBP–CBD-cellulose can be desorbed or released, and whether the regenerated PdBP–CBD-cellulose can be reused in a Pd(II) removal experiment. As shown in Fig. 2C, the PdBP–CBD was regenerated after the bound Pd(II) was removed through washing. The high removal efficiency was maintained for at least three binding/removal (regeneration) cycles involving the treatment with 1 M thiourea. The capability of PdBP–CBD to retain its reusability may be explained by the fact that palladium binding peptides were obtained from peptide libraries through multiple binding and washing cycles.10 As a result, the obtained PdBP is able to specifically bind to palladium and retain its desorption and adsorption performance after several cycles.
Adsorption kinetics
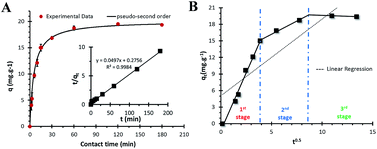
Fig. 3A shows the effect of contact time on adsorption. The results showed that the adsorption capacity reached equilibrium at approximately 60 min. The adsorption efficiency increased gradually at first and then reached equilibrium as time passed. This trend suggested that the adsorption sites were void in the beginning; thus, adsorbates could easily interact with these sites during the initial stage, but after a lapse in time the remaining vacant sites became more difficult to be occupied by the adsorbates.Studying kinetics is a crucial step for understanding the adsorption mechanism and for designing columns in laboratory, pilot, and industrial scales.21 Several models can be used to express the mechanism of sorption onto a sorbent. In this study, Lagergren pseudo-first order22 and pseudo-second order models,23 and intraparticle diffusion models24 were used to evaluate the mechanism of Pd(II) adsorption onto PdBP–CBD-cellulose. The linear form of the Lagergren pseudo-first order model is expressed as eqn (3)
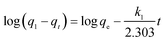 | (3) |
The linear form of pseudo-second order equation can be written as eqn (4):
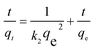 | (4) |
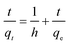 | (5) |
The intraparticle diffusion kinetic model can be written as eqn (6)
| qt = kidt0.5 + Ci | (6) |
As shown in Table 1, the correlation coefficient values (R2) of the pseudo-second order model (Fig. 3A, inset) were slightly higher than those of the pseudo-first order and intraparticle diffusion models (Fig. 3B). The intercept, which did not equal q1 showed that the reaction is not likely to be a first order reaction. The calculated adsorption capacity (qe,cal) values from pseudo-second order model were close to experimental adsorption capacity (qe,exp) values, indicating that the pseudo-second order kinetic model is suitable for modelling the adsorption of Pd(II).
| Model | Parameter | Pd(II) |
|---|---|---|
| Pseudofirst-order kinetic | k1 (min−1) | 0.0252 |
| qe,cal (mg g−1) | 10.23 | |
| R2 | 0.8186 | |
| Pseudosecond-order kinetic | k2 (g mg−1 min−1) | 0.0100 |
| h (mg g−1 min−1) | 4.0519 | |
| qe,cal (mg g−1) | 20.1613 | |
| R2 | 0.9980 | |
| Intraparticle diffusion | kid (mg g−1 min−0.5) | 1.3985 |
| R2 | 0.7394 |
To elucidate the diffusion mechanism and investigate whether intraparticle diffusion is rate-limiting factor in the adsorption process, the intraparticle diffusion model (eqn (6)) was considered. Fig. 3B presents an intraparticle diffusion plot for Pd(II). If intraparticle diffusion is a rate-limiting factor, then plots of qt versus t0.5 show a linear relationship. The slope of the linearized plot illustrates the rate parameter of intraparticle diffusion, and the intercept is proportional to the thickness of the boundary layer (film). A value of constant Ci in the intraparticle diffusion equation equal to zero, indicates that intraparticle diffusion governs the adsorption rate of the entire adsorption process. In general, the plot of qt against t0.5 usually shows more than one linear portion; when the slope of the first portion is not zero, film diffusion is determined to control the adsorption rate at the beginning.26
As shown in Fig. 3B, the linear fit of the intraparticle diffusion model showed three linear portions, indicating that the adsorption process consists of multiple stages. Each stage was identified according to a change in the slope of the linear line which was used to fit the experimental data. The first stage was assumed to be caused by external surface adsorption or instantaneous adsorption which is driven by initial differences in the metal ions concentrations. The second linear stage is the gradual adsorption stage, in which intraparticle diffusion is the rate-limiting step. The third stage is the final equilibrium stage, in which intraparticle diffusion slows because of the low concentration of residual metal ions in the solution. As shown in Fig. 3B, the plots were not linear throughout the adsorption process, implying that more than one process affected the adsorption.
Adsorption isotherms
To evaluate the adsorption capacity of PdBP–CBD-cellulose, the adsorption isotherm of Pd(II) was studied by varying the initial concentration of the solution while keeping all other parameters constant. The Langmuir and Freundlich isotherm equations were used to interpret the adsorption experimental data. The Langmuir isotherm model, which is based on monolayer adsorption onto the active sites of the adsorbent, is used to evaluate adsorption systems. The expression of the Langmuir model is as follows:
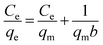 | (7) |
The Freundlich isotherm assumes heterogeneous adsorption because of the diversity of the adsorption sites or the diverse nature of the metal ions adsorbed. The Freundlich adsorption equation is expressed as:
| qe = kC1/ne | (8) |
The Pd(II) adsorption profile of PdBP–CBD-cellulose and the adsorption isotherms parameters for Pd(II) onto PdBP–CBD-cellulose is shown in Fig. 4. The values of regression coefficients obtained from the models were used as fitting criteria to identify these isotherms. Under the experimental conditions, the Langmuir model showed a fit (R2 > 0.98) that was superior to that of the Freundlich isotherm models, suggesting that Pd(II) adsorption on PdBP–CBD-cellulose is based on monolayer coverage. According to the Langmuir isotherm, the maximum adsorption capacity qm of the adsorption of Pd(II) on PdBP–CBD-cellulose was 175.44 mg g−1. Interestingly, a back-of-the-envelope calculation suggested that the moles of palladium bound at capacity is in excess of the moles of PdBP protein present. Hypothetically this might be attribute to two reasons. The first one is that each binding protein has more than one binding site. The Langmuir isotherm is based on the interaction between adsorbates and the binding sites, instead of the binding molecules. Therefore, this result suggested that more than one binding site are presented in each PdBP protein. Second, the Pd(II) ions somehow forms nanoparticles on the adsorbent, resulting in a higher adsorption capacity. Since this peptide has been used for nano-palladium synthesis,10 it is not surprised that nanoparticles are formed on the peptides. For the Freundlich isotherm, the n value was between 1 and 10, suggesting that adsorption was favourable in the experimental conditions.
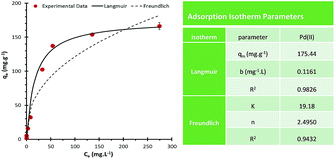 | ||
| Fig. 4 Adsorption isotherms of Pd(II) on PdBP–CBD-cellulose (100 mg cellulose, 1.73 mg PdBP–CBD, 1.5 mL Pd(II) solutions, pH 3.54 ± 0.2, 3 h, 25 °C). | ||
Lemna minor growth inhibition test
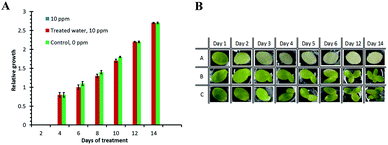
Rain can transport palladium deposited near roads into aquatic environments, resulting in the contamination of aquatic ecosystems. The solubility of palladium from spent automotive catalysts in water is relatively high, suggesting that some of the palladium is not in metal forms but in soluble forms, such as chloride.27 Palladium was reported to have the highest bioavailability among the PGEs.28 Consequently, substantial amounts of palladium are consumed by plants and aquatic organisms and this can be harmful to human health. To evaluate the exposure of natural ecosystems to palladium, environmental indicators (bioindicators) have been used as a direct or indirect measure of environmental quality. In this study, we used L. minor as a bioindicator and PdBP–CBD-cellulose to explore the possibility of palladium bioremediation. The rapid growth of L. minor and the susceptibility of L. minor to palladium made L. minor the most suitable palladium environmental bioindicator. L. minor fronds were observed daily for toxicity symptoms (chlorosis and frond dislocation).Fig. 5A shows the relative growth rates of plants in samples over 14 days of treatment. On the second day, no growth was observed in all samples. On the fourth day, significant differences in RG were observed among the samples. The plants in samples of 10 ppm palladium (A) showed no growth (100% growth inhibition). Samples of palladium-treated water (B) showed no significant difference with respect to the control (C). Throughout the treatment, the plants in samples of 10 ppm palladium (A) kept 100% growth inhibition whereas those of treated water (B) maintained their relative growth rates with respect to the control (C).
Fig. 5B shows images of fronds grown in different samples over 14 days of treatment. On the first day, no significant differences between the tested samples (A) and (B) and the control (C) were observed. On the second day, the plants in A-group samples exhibited loss of green pigments (chlorosis) compared with the plants in the (B) and (C) groups. In the subsequent days, the plants in the A samples showed significant chlorosis while no colour changes were observed in the plants in the (B) and (C) groups. Fig. 5A shows that there was no dislocation of fronds or loss of green pigment in both samples (B) and (C) after 14 days of treatment; these healthy L. minor samples in treated water indicated that bioremediation was effective.
Conclusions
In this study, we created a biosorbent for palladium recovery and detoxification. This strategy involved taking advantage of (1) cellulose, which is naturally abundant, inexpensive, and chemically inert; (2) CBDs, which bind rapidly with cellulose; and (3) PdBPs, which bind selectively with palladium ions. By genetically fusing PdBPs with CBDs and mechanically mixing PdBP–CBD with cellulose, we creates a reusable and highly selective biosorbent for recovering palladium ions. In addition, the biomolecule-cellulose-based adsorbent can be used as an effective, renewable, and environmentally friendly remediation method for metal detoxification.Acknowledgements
This study was financially supported by the Ministry of Science and Technology of the Republic of China, Taiwan under the Contract no. NSC 101-2218-E-011-046-MY3. The authors are grateful to Prof. Wilfred Chen at University of Delaware for the plasmid pCBD-Blue.Notes and references
- F. Zereini, F. Alt, J. Messerschmidt and A. von Bohlen, et al., Environ. Sci. Technol., 2004, 38, 1686 CrossRef CAS.
- I. S. Sen, Environ. Sci. Technol., 2013, 47, 13903 CrossRef CAS PubMed.
- S. De Corte, T. Hennebel, B. De Gusseme and W. Verstraete, et al., Microb. Biotechnol., 2012, 5, 5 CrossRef CAS PubMed.
- N. Das, Hydrometallurgy, 2010, 103, 180 CrossRef CAS PubMed.
- E. Guibal, Sep. Purif. Technol., 2004, 38, 43 CrossRef CAS PubMed; A. Ramesh, H. Hasegawa, W. Sugimoto and T. Maki, et al., Bioresour. Technol., 2008, 99, 3801 CrossRef PubMed; L. Zhou, J. Xu, X. Liang and Z. Liu, J. Hazard. Mater., 2010, 182, 518 CrossRef PubMed.
- D. Parajuli, K. Inoue, H. Kawakita and K. Ohto, et al., Miner. Eng., 2008, 21, 61 CrossRef CAS PubMed.
- M. Gurung, B. B. Adhikari, S. Alam and H. Kawakita, et al., Chem. Eng. J., 2013, 228, 405 CrossRef CAS PubMed.
- P. Yong, N. A. Rowson, J. P. G. Farr and I. R. Harris, et al., J. Chem. Technol. Biotechnol., 2002, 77, 593 CrossRef CAS; I. De Vargas, L. E. Macaskie and E. Guibal, J. Chem. Technol. Biotechnol., 2004, 79, 49 CrossRef.
- S. W. Won, P. Kotte, W. Wei and A. Lim, et al., Bioresour. Technol., 2014, 160, 203 CrossRef CAS PubMed; L. Liu, S. Liu, Q. Zhang, C. Li and C. Bao, et al., J. Chem. Eng. Data, 2012, 58, 209 CrossRef; M. Can, E. Bulut and M. Özacar, J. Chem. Eng. Data, 2012, 57, 2710 CrossRef.
- U. O. S. Seker and H. V. Demir, Molecules, 2011, 16, 1426 CrossRef CAS PubMed; M. Sarikaya, C. Tamerler, A. K. Y. Jen and K. Schulten, et al., Nat. Mater., 2003, 2, 577 CrossRef PubMed; D. B. Pacardo, M. Sethi, S. E. Jones and R. R. Naik, et al., ACS Nano, 2009, 3, 1288 CrossRef PubMed.
- I. Levy and O. Shoseyov, Biotechnol. Adv., 2002, 20, 191 CrossRef CAS.
- A. A. Wang, A. Mulchandani and W. Chen, Biotechnol. Progr., 2001, 17, 407 CrossRef CAS PubMed; M. Linder, T. Nevanen, L. Söderholm and O. Bengs, et al., Biotechnol. Bioeng., 1998, 60, 642 CrossRef; T. Y. Shih and S. L. Tsai, RSC Adv., 2014, 4, 40994 RSC.
- OECD, OECD Publishing, 2009 Search PubMed.
- H. E. Ensley, J. T. Barber, M. A. Polito and A. I. Oliver, Environ. Toxicol. Chem., 1994, 13, 325 CrossRef CAS.
- H. Wang, C. Li, C. Bao and L. Liu, et al., J. Chem. Eng. Data, 2011, 56, 4203 CrossRef CAS; K. Fujiwara, A. Ramesh, T. Maki and H. Hasegawa, et al., J. Hazard. Mater., 2007, 146, 39 CrossRef PubMed.
- M. Can, E. Bulut, A. Örnek and M. Özacar, Chem. Eng. J., 2013, 221, 146 CrossRef CAS PubMed; I. Puigdomenech, Hydra and Medusa, Version, 2010, http://www.kemi.kth.se/medusa/.
- L. Zhou, J. Liu and Z. Liu, J. Hazard. Mater., 2009, 172, 439 CrossRef CAS PubMed; P. Chassary, T. Vincent, J. S. Marcano and L. E. Macaskie, et al., Hydrometallurgy, 2005, 76, 131 CrossRef PubMed.
- D. S. Moore, Biochem. Educ., 1985, 13, 10 CrossRef CAS.
- C. Mack, B. Wilhelmi, J. R. Duncan and J. E. Burgess, Biotechnol. Adv., 2007, 25, 264 CrossRef CAS PubMed.
- T. P. Nguyen, N. P. Hankins and N. Hilal, Desalination, 2007, 204, 277 CrossRef CAS PubMed.
- I. Ali and V. K. Gupta, Nat. Protoc., 2006, 1, 2661 CrossRef CAS PubMed.
- Y. S. Ho, Scientometrics, 2004, 59, 171 CrossRef CAS.
- Y. S. Ho and G. McKay, Process Biochem., 1999, 34, 451 CrossRef CAS.
- J. W. Weber and J. C. Morris, J. Sanit. Eng. Div. Am. Soc. Civ. Eng., 1963, 89, 31 Search PubMed.
- Y. S. Ho and G. McKay, Chem. Eng. J., 1998, 70, 115 CrossRef CAS.
- J. Zhou, S. Yang, J. Yu and Z. Shu, J. Hazard. Mater., 2011, 192, 1114 CrossRef CAS PubMed.
- K. E. Jarvis, S. J. Parry and J. M. Piper, Environ. Sci. Technol., 2001, 35, 1031 CrossRef CAS.
- M. Moldovan, S. Rauch, M. Gómez and M. Antonia Palacios, et al., Water Res., 2001, 35, 4175 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Details of primers used in this study; plasmid construction; pH test of Pd–CBD binding to microcrystalline cellulose; and the distribution of palladium species based on Hydra and Medusa computer program. See DOI: 10.1039/c4ra16200e |
| This journal is © The Royal Society of Chemistry 2015 |