Abietane diterpenoid of Salvia sahendica Boiss and Buhse potently inhibits MCF-7 breast carcinoma cells by suppression of the PI3K/AKT pathway†
Abstract
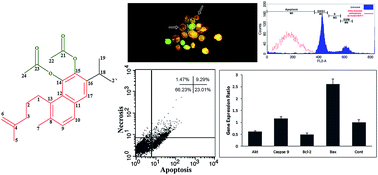
In the current study, we report on the bioactive compounds isolated from the roots of Salvia sahendica Boiss and Buhse using bioassay-guided procedures and their biological effects against MCF-7 breast carcinoma cells. In comparison with other solvents, the hexane-based extraction resulted in the most potent anti-cancer activity, and hence it was subjected to more phytochemical fractionation analyses using vacuum liquid chromatography (VLC), reversed-phase high pressure liquid chromatography (HPLC) and NMR spectroscopy. The biological impacts of the isolated pure compounds were evaluated using MTT, DAPI and acridine orange/ethidium bromide staining (AO/EB) assays. Cell cycle analysis was performed to assess the sub-G1 population of the cells treated with the extracted compounds, while the FITC-labeled annexin V assay was used to study the apoptosis profile. The gene expression profile of the treated cells was studied by quantitative PCR, looking at key genes (Caspase 9, Bax, Akt and Bcl-2) involved in apoptosis. Ketoethiopinone (1) and ortho-diacetate aethiopinone (2) compounds were identified using 1H and 13C-NMR. Compounds 1 and 2 showed profound inhibitory impact on the treated MCF-7 cells with IC50 values of 8.6 and 14.2 μg mL−1 at 48 h, respectively. DAPI and AO/EB assays resulted in significant alterations in the nucleus through chromatin remodeling in the treated cells which somewhat impacts the integrity of the cell membrane. An annexin V flow cytometry assay revealed that the cells treated with compound 2 resulted in early and late apoptosis (∼30%). Gene expression profiling demonstrated significant (p < 0.05) changes in the expression of Bcl-2, Caspase 9, Bax and Akt in the cell treated with compound 2 with profound impact on the Bax and Akt pathways. Taken all together, we propose ortho-diacetate aethiopinone as a new class of anticancer agents with great translational potential for clinical uses against solid tumors.

 Please wait while we load your content...
Please wait while we load your content...