Synthesis, characterization and enhanced bactericidal action of a chitosan supported core–shell copper–silver nanoparticle composite†
Sadhucharan Mallicka,
Pallab Sanpuia,
Siddhartha Sankar Ghoshbc,
Arun Chattopadhyayac and
Anumita Paul*a
aDepartment of Chemistry, Indian Institute of Technology Guwahati, Guwahati 781039, India. E-mail: arun@iitg.ernet.in; anumita@iitg.ernet.in; Fax: +91 3612582349; Tel: +91 3612582304 Tel: +91 3612582308
bDepartment of Biotechnology, Indian Institute of Technology Guwahati, Guwahati 781039, India
cCentre for Nanotechnology, Indian Institute of Technology Guwahati, Guwahati 781039, India
First published on 9th January 2015
Abstract
A simple two-step seed-mediated method has been developed to synthesize chitosan (CS)-supported core–shell metal nanoparticles (NPs) in aqueous solution. CS-supported copper NPs (CS–Cu NPs), when treated with silver nitrate (AgNO3) solution, resulted in Ag shells on Cu NP cores in the composite. The Cu@Ag NPs in the CS composite were characterized and found to be spherical in shape with an average particle size of 14.0 ± 3.4 nm. The antibacterial efficacy of the CS–Cu@Ag NP composite was evaluated on Gram-negative Escherichia coli and Gram-positive Bacillus cereus bacteria using turbidity measurement as well as flow-cytometry. Results demonstrated enhanced bactericidal activity of the composite. Analytical techniques, used to elucidate the mode of bactericidal action, revealed that the superior antibacterial activity of the CS–Cu@Ag NP composite was due to synergistic effects of CS and Cu@Ag NPs. The proposed mode of antibacterial action involves the electrostatic binding of CS to the bacterial cell wall and simultaneous interaction of Cu@Ag NPs with membrane proteins leading to perforation of the cell membrane and release of intracellular material.
1. Introduction
The bactericidal properties of silver and copper have been well established since ancient times. Moreover, silver nanoparticles (Ag NPs) and copper nanoparticles (Cu NPs) have potential bactericidal properties, either individually or in polymer composites.1–5 In our previous studies, we have prepared iodinated chitosan (CS)–Ag NP and iodinated CS–Cu NP composites and investigated their bactericidal activity on Gram-negative green fluorescent protein (GFP) expressing recombinant Escherichia coli (E. coli) and Gram-positive Bacillus cereus (B. cereus) bacteria.1,2 Whereas CS is a natural biopolymer having bactericidal properties, Ag and Cu are elements with well-known antibacterial characteristics. However, Ag and Cu are cytotoxic at doses greater than 0.1 mg L−1 and 1.3 mg L−1, respectively, in drinking water according to U.S. Environmental Protection Agency reports.6 Ag, in particular, accumulates in the human body and thus can potentially reach toxic levels from sustained environmental exposures. On the other hand, Cu does not accumulate in the human body due to a sequestering mechanism which helps its excretion from the body.7,8 Hence Cu NPs are a better alternative to Ag NPs in biological applications, except for the lower stability of Cu NPs under ambient atmospheric conditions. Freshly prepared Cu NPs, within hours of synthesis, formed copper oxide on the surface of Cu NPs, which increased their minimal inhibitory concentration (MIC) value against Gram-negative bacteria E. coli, thereby reducing their antibacterial properties.2,9 Therefore, stabilizing Cu NPs against oxidation and harnessing maximal bactericidal efficacy is challenging. One possible way of circumventing this problem could be through Ag shell formation on the Cu core, i.e. through the use of Cu@Ag core–shell NPs, instead of pure Ag NPs or pure Cu NPs.Bimetallic NPs consisting of two different metals offer greater advantages over their monometallic counterparts, since their optical, catalytic and biological properties can be tuned by changing the composition.10–21 Bimetallic colloids are of two types, viz., alloy and core–shell. Commonly used methods of synthesizing Cu@Ag NPs include vacuum vapour deposition,13,22 sono-chemical reduction,23 two-step polyol reduction,14,24 sequential ion implantation,25 sequential ion-exchange,26,27 electroless plating28 and galvanic replacement reaction.12 But these synthetic methods are harsh and not suitable for biological applications. Moreover, great care is to be taken in using oxygen free environment to prevent aerial oxidation of the Cu NPs. For example, Langlois et al. used ultra-high vacuum technique to obtain Cu@Ag bimetallic nanoparticles on a substrate by sequential deposition22 and Tsuji et al. synthesized Cu@Ag NPs using a polyol reduction method under bubbling of N2 gas.14,24
Bimetallic Cu@Ag NPs are reported to have improved resistance towards oxidation as compared to pristine Cu NPs.29,30 Core–shell NPs have been used for catalytic, optical and magnetic applications. In catalytic reactions, the association of two different metals offers superior activity, increased stability and better selectivity.31–33 Enhanced antibacterial activity of core–shell nanostructures has also been reported.34–38 Bactericidal core–shell NPs consisting of Ag shell supported on organic and inorganic cores have been studied. However, the bactericidal efficacy of core–shell NPs consisting of Ag shell and Cu as core has not been studied, despite the fact that Cu has well known antibacterial and antifungal properties.2 Recently, several groups have reported that Cu–Ag alloy NPs display superior bactericidal activity against E. coli as compared to pure Ag NPs.39,40 Individually, elemental Cu is less-toxic to humans but substantially toxic to microorganisms.41 On the other hand, Ag NPs are more effective because both Ag ions and Ag NPs have excellent biocidal ability.1,3–5,42,43 One of the ways of reducing the effects of toxicity of Ag NPs to human is by replacing the core of the Ag NPs by a less-toxic, yet antimicrobial metal, such as Cu.
Here, we report a simple two-step seed-mediated route to prepare CS-supported Cu core–Ag shell NPs (CS–Cu@Ag NPs) with enhanced bactericidal properties compared to individual Cu NPs and Ag NPs. We have recently reported the synthesis of CS-supported Cu NPs using hydrazine hydrate as a reducing agent.2 In this work, Cu NPs were prepared in a similar fashion and used as seed particles over which Ag+ ions were reduced by transmetalation reaction to generate the Cu@Ag NPs. The resulting CS–Cu@Ag NP composite was stable for several weeks and was able to significantly retard bacterial growth at a very low Ag concentration.
2. Experimental
2.1 Materials and methods
Chitosan of high molecular weight (75% deacetylated; Sigma-Aldrich Chemical Co., India), copper(II) sulphate pentahydrate (CuSO4·5H2O; Merck, India), silver nitrate (AgNO3, 99.5%; Merck, India), hydrazine hydrate (80% solution, Merck, India), sodium hydroxide (NaOH, 98%; Merck, India) and acetic acid (glacial, 99–100%; Merck, India) were used as obtained without any additional purification. Milli-Q grade water was used throughout the experiments. Luria–Bertani (LB) broth and nutrient broth (NB) were procured from HiMedia, India. Molecular grade agarose for gel-electrophoresis was purchased from Sigma-Aldrich Chemical Co., India. GFP-expressing recombinant E. coli (MTCC433)3 and B. cereus (MTCC1305) were grown at 37 °C for 12 h in LB and NB media, respectively.2.2 Synthesis of core–shell CS–Cu@Ag NP composite
Seed-mediated growth method was employed to prepare CS–Cu@Ag NPs. The Cu NP seeds were prepared by a recently published method wherein the Cu NPs were synthesized by reduction of CuSO4 with hydrazine hydrate in the presence of CS as stabilizer.2 To this freshly prepared 30 mL Cu NP seed dispersion, 0.6 mL (amount optimized based on preliminary studies, Fig. S5, ESI†) of 0.02 M AgNO3 solution was injected at room temperature and stirred for 12 h. The obtained orange reddish solution was centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, the pellet washed with Milli-Q water and re-dispersed in 30 mL of water that contains 0.25% (v/v) acetic acid. Prior to bactericidal experiments, the pH of that composite solution was set to 6.3 for the better dispersion of the CS–Cu@Ag NP composite in the bacterial medium as reported previously.4
000 rpm, the pellet washed with Milli-Q water and re-dispersed in 30 mL of water that contains 0.25% (v/v) acetic acid. Prior to bactericidal experiments, the pH of that composite solution was set to 6.3 for the better dispersion of the CS–Cu@Ag NP composite in the bacterial medium as reported previously.4
2.3 Characterization of core–shell CS–Cu@Ag NP composite
UV-visible spectra of the NP dispersions were recorded using a Hitachi U-2900 Spectrophotometer. Transmission electron microscopic (TEM) and high resolution TEM (HRTEM) imaging were carried out in a JEM-2100 (JEOL, Japan) instrument at an accelerating voltage of 200 keV. Five microliters of dispersed solution was drop-cast onto TEM grids coated with a thin carbon film, air-dried and then the samples analyzed under the TEM. Scanning electron microscopy (SEM; LEO 1430VP, Germany) coupled with energy dispersive X-ray spectroscopy (EDX) and field emission SEM (FESEM, Carl Zeiss, SIGMA VP) coupled with EDX instrument were used for the study of surface morphology and elemental analysis of CS–Cu@Ag NP composite and composite treated bacteria. Ten microliters of a dispersed sample was placed on an Al foil-wrapped glass slide, allowed to air dry and finally sputter-coated with Au using a sputter coater (SC7620-MiniPolaron Sputter Coater, Quorum Technologies, England), before analysis. Powder X-ray diffraction (XRD) experiments were carried out using Philips diffractometer (Model 1715) with Cu Kα1 radiation (λ = 1.54060 Å), operating at 55 kV and 250 mA. Zeta potential (ζ) values were determined using Delsa™ Nano Submicron Particle Size and Zeta Potential Particle Analyzer (PN A54412AA, Beckman Coulter). Fourier transform infrared (FTIR) spectroscopic characterization were carried out using a Perkin Elmer spectrum one spectrometer for chemical analysis of solid CS and solid CS–Cu@Ag NPs by forming pellets with KBr. X-ray photoelectron spectroscopy (XPS) measurements were performed to characterize the core–shell structure of the CS–Cu@Ag NP composites. For XPS measurements, powdered CS–Cu@Ag NP composite was pressed into 0.5 mm thick and 6.0 mm diameter pellet and placed in an ultra-high vacuum chamber (1 × 10−10 Torr) of a PHI 5000 Versa Probe II (ULVAC-PHI, INC, Japan) equipment employing Al Kα X-rays (hν = 1486.6 eV). A combination of low energy Ar+ ions and electrons was used for charge neutralization in each measurement. The binding energy (eV) was corrected against C 1s (284.6 eV) reference standard. Elemental composition in XPS spectra was acquired with analyzer pass energy of 187.85 eV, while high-resolution XPS spectra were acquired with analyzer pass energy of 58.70 eV with step size of 0.125 eV at 50 ms per step. Quantitative estimation of Cu and Ag present in CS–Cu@Ag NP composite was made using an atomic absorption spectrophotometer (AAS) (Model: AA240 Varian Inc) following dissolution of the composite in dilute HCl solution. In order to determine the amount of Cu and Ag ions discarded after synthesis of the composite, the supernatant collected after centrifugation of the reaction mixture was also subjected to AAS measurement.2.4 Bactericidal studies
For antibacterial experiments, GFP-expressing E. coli (Gram-negative bacteria) and B. cereus (Gram-positive bacteria) were grown in LB ampicillin and NB media, respectively. Bacteria (108 cfu mL−1) were grown in media in the presence of different concentrations of CS–Cu@Ag NP composite for 12 h at 37 °C. The lowest concentration of composite at which there was no visual turbidity was taken as the MIC value of the composite. Further, the cultures which lacked visual turbidity were re-inoculated in fresh media. The lowest concentration of the composite that killed at least 99.9% of original inoculum and resulted in no visual turbidity after re-inoculation was taken as minimum bactericidal concentration (MBC). The experiments were performed at least in triplicate to ensure reproducibility. The growth of bacteria in a sample was monitored at different times by recording the optical density (OD) at 595 nm using a Perkin Elmer Lambda 45 Spectrophotometer.2.5 Flow-cytometric analysis for bacterial cell viability
Disruption of membrane integrity and cell viability of GFP-expressing E. coli bacteria in presence of CS–Cu@Ag NP composite were assessed using the nucleic acid dye, propidium iodide (PI), in addition to the constitutively expressed GFP. As a membrane impermeable dye, PI cannot bind with the DNA of viable cells. However, PI permeates into dead cells, with compromised cell-membrane, binds DNA and exhibits fluorescence at 620 nm when excited by the 488 nm blue laser.1,2 Briefly, 500 μL (108 cells per mL) of recombinant E. coli cells and 250 μL of the CS–Cu@Ag NP composite (at MIC of 63.4 μg mL−1) were incubated together for various time periods (2, 4, 6 h) at 25 °C. Similar experiments were carried out with 500 μL (108 cells per mL) of the recombinant E. coli cells and 375 μL of the composite (at MBC of 92.99 μg mL−1). Following incubation, 1.5 μL of PI (0.2 mM) was added to every tube and diluted with 150 mM solution of sodium chloride. Flow cytometric measurements were performed on a BD FACSCalibur System, (BD Biosciences, CA, USA), where the samples were irradiated with a 15 mW argon ion laser (488 nm), and fluorescence detected through standard configuration of 525 ± 10 nm (green) and 620 ± 10 nm (red) band pass filters. Fluorescence of different cell populations in dot plots, i.e. live, compromised, dead and lysed, was gated based on the different viability stages of the cells. This method permits the separation of different cell populations according to membrane integrity.442.6 DNA isolation and agarose gel electrophoresis
Recombinant GFP plasmid (pGFP) was isolated from E. coli (control) and from CS–Cu@Ag NPs treated E. coli cells by alkaline lysis method. Plasmids were then run in an agarose gel and subsequently stained with ethidium bromide (EtBr) for visualization under UV-transilluminator.45,462.7 Cytotoxicity assay
HEK-293 (human embryonic kidney) cell line was maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U mL−1 penicillin and 50 mg mL−1 streptomycin in a 5% CO2-containing humidified atmosphere at 37 °C. The toxicity of CS–Cu@Ag NP composite on HEK-293 cells was determined by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. For this, HEK-293 cells were seeded into a 96-well plate (5 × 103 cells per well) and grown overnight. Following treatment with MIC and 2× MIC of the composite for 24 h and 48 h, MTT (1 mg mL−1) was added to each well and incubated for 3 h. The formed formazan product was solubilized in DMSO and absorbance measured at 550 nm in a microplate reader (Bio-Rad model 680; Bio-Rad, CA).3. Results and discussion
3.1 Synthesis and characterization of CS–Cu@Ag NP composite
The CS-stabilized Cu seed NPs were synthesized under ambient condition by reacting alkaline CuSO4 solution with hydrazine hydrate and CS, based on a procedure developed by our group previously.2 The reddish precipitate so-obtained was isolated from the reaction medium and re-dispersed immediately in water acidified with acetic acid (0.25%, v/v). The resulting solution showed an absorption peak at 583 nm in the UV-vis spectrum indicating the formation of Cu NPs. This was further confirmed by TEM analysis (refer to ESI, Fig. S1†).2 When AgNO3 solution was added to the reddish CS–Cu NP dispersion, the colour changed to orange-red due to the formation of CS–Cu@Ag NP composite. The orange-red dispersion was centrifuged, the pellet re-dispersed in Milli-Q water and its UV-vis spectrum recorded. This orange-red solution was found to strongly absorb at 417 nm (Fig. 1) which is attributed to the surface plasmon resonance (SPR) band of Ag NPs. This dispersion was subjected to further characterization and also employed for bactericidal studies.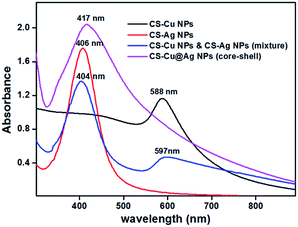 | ||
| Fig. 1 UV-visible spectra of CS–Cu@Ag NPs (pink line), CS–Cu NPs seed particles (black line), CS–Ag NPs alone (red line), mixture of CS–Cu NPs and CS–Ag NPs (blue line). | ||
In the present method, Ag+ ions from AgNO3 were reduced to Ag(0) by the Cu(0) atoms on the surface of Cu NPs. In other words, galvanic replacement of Cu atoms by Ag atoms on the surface of the Cu NPs led to the formation of Ag shell over the Cu core.12 After centrifugation of the orange-reddish dispersion, AAS measurements of the supernatant revealed that the concentration of Cu2+ ions typically increased from 1.774 μg mL−1 to 0.2136 mg mL−1 upon addition of 3.77 mg mL−1 of Ag+, whose concentration in turn dropped to 1.355 μg mL−1 in the supernatant. These values are consistent with a galvanic replacement reaction of Cu0 to Cu2+ which was released into the solution, while Ag+ was simultaneously taken up from the solution to be deposited as Ag0 on the surface of the Cu NPs.
The CS–Cu@Ag NP composite showed a single broad peak around 417 nm. When CS–Cu NPs and CS–Ag NPs were prepared independently and mixed subsequently, UV-vis spectrum showed two distinct SPR bands: one at 599 nm for Cu NPs and another at 405 nm for Ag NPs (Fig. 1). It is reported that the SPR band for Ag–Cu alloy NPs appears between the SPR band for pure Cu NPs (560–600 nm) and that of pure Ag NPs (390–420 nm), with the peak position of the SPR band shifting to longer wavelengths with increasing Cu percentage.39,40,47 Thus the present composite showing a peak at 417 nm, neither consisted of Cu–Ag alloy NPs nor consisted of a mixture of Ag NPs and Cu NPs. The band at ∼417 nm corresponds to SPR of Ag NPs. Moreover, there was no SPR band observed due to Cu NPs. Based on the above set of observations, it is likely that our composite contained Cu@Ag NPs with Ag as the shell. The dampening of SPR band for a core metal in presence of a shell metal has already been reported for Cu@Ag NP having an outer Ag shell of thickness greater than 5–7 nm.26,27
Furthermore, the formation of CS–Cu@Ag NPs was clearly evident in TEM images (Fig. 2a) which showed the presence of almost spherical particles with an average size of 14.0 ± 3.4 nm. Closer inspection of the TEM images (Fig. 2a and b) revealed that most of the particles had a darker central part (core) and lighter outer part (shell). The average shell thickness was ∼5.8 nm while the average core diameter was ∼9.5 nm. HRTEM image of a single particle (Fig. 2b inset) and corresponding selected area electron diffraction (SAED) pattern (Fig. 2c) clearly indicated crystalline nature of the Cu@Ag NPs. In particular, the HRTEM image in Fig. 2b showed the presence of lattice fringes which were further enhanced by inverse fast Fourier transform analysis (IFFT) and shown in Fig. 2d and e. A lattice fringe spacing of 0.196 nm for the core metal and 0.238 nm for the outer shell metal was clearly visible in the IFFT images (Fig. 2d and e). The lattice spacing of 0.196 nm corresponds to Cu(111) plane,2 while that of 0.238 nm corresponds to Ag(111) plane.1,48 These observations indicated that the dark central part was due to metallic Cu and lighter outer part due to metallic Ag.
Similar conclusions can be drawn from the SAED pattern shown in Fig. 2c, where the diffraction rings were indexed. The indices are listed in Table S1 (refer to ESI†). The EDX analysis carried out during FESEM (Fig. S2, refer to ESI†) also showed the presence of both the elements Cu and Ag, as expected for the core shell structure of the CS–Cu@Ag NP composite. Our observations shown in Fig. 2 and S2,† undoubtedly supported the presence of Cu@Ag NPs in the CS polymer matrix. Cu and Ag metal content in the CS-based composite were determined by AAS measurements. Alternatively, the difference in the amount of Cu2+ and Ag+ ions in the initial feed solution to their amounts being discarded in supernatant solution allowed indirect estimation of the Cu2+ ions and Ag+ ions taken up by the CS–Cu@Ag NP composite. The amount of Cu and Ag content in the composite so measured by the two methods were found to be similar within the experimental error.
XRD pattern of a typical sample of CS–Cu@Ag NP composite recorded after 24 h of synthesis is shown in Fig. 2f. The diffraction peaks at 2θ = 38.2, 44.3, 64.6, 77.4, and 81.4 are due to reflections from (111), (200), (220), (331), (222) planes of metallic Ag (JCPDS-04-0783), respectively, adopting a face-centred-cubic lattice structure (Fig. 2f and S3a, ESI†). It is noteworthy that the XRD pattern in Fig. 2f lacks peaks due to the pure Cu, indicating that Cu is well-shielded by the Ag shell in the CS–Cu@Ag NP composite. In most metallic core–shell NP systems, the XRD peaks from the core metal are not observed due to the core metal being in kinematic diffraction state.23,26,48–50 Before the formation of Ag shell in the present method, however, the CS–Cu NP sample showed distinct peaks at 2θ values of 43.4°, 50.5° and 74.2° corresponding to diffraction from (111), (200), and (220) planes of Cu(0), respectively (refer to ESI, Fig. S3b†).
The orange-red CS–Cu@Ag NP dispersion was stable for several weeks. CS, a naturally existing cationic polysaccharide, is able to chemisorb on Cu surface due to the presence of functional groups i.e. primary amines (–NH2) and hydroxyl (–OH) groups.1,2 Hence, CS–Cu NP seed particle showed slower rate of oxidation than the free NPs.2 FTIR analysis confirmed the presence of CS in the nanocomposite material (Fig. S4, refer to ESI†). Additionally, the UV-visible spectra, XRD pattern and TEM images of aged CS–Cu@Ag NPs showed that the samples were stable in the medium for more than two weeks under ambient conditions (refer to ESI, Fig. S5†). This is to be contrasted with the lower stability of CS–Cu NP samples (typically 12 h) reported in our earlier studies.2
Chemical composition near the surface of CS–Cu@Ag NP composite was investigated by XPS, where the wide energy scan revealed the presence of elemental Ag, Cu, O, N, and C in the sample (refer to ESI, Fig. S6†). The corresponding high-resolution XPS result for Cu and Ag element is shown in Fig. 3a and c, respectively. The binding energy for Cu 2p3/2 and Cu 2p1/2 showed two peaks at 932.4 eV and 952.2 eV, associated with Cu(0) electrons. In addition, well known shake-up satellite peaks appeared at 939 eV and 942 eV in Cu 2p spectrum, indicating the presence of CuOx species (Fig. 3a).39,51 This trace CuO formation on the surface regions of CS–Cu@Ag NP sample indicated that at least some Cu NPs were not sufficiently covered by the Ag shell, making them prone to aerial oxidation.
 | ||
| Fig. 3 High-resolution XPS spectra of Cu 2p region (a) before and (b) after sputtering ∼10 nm depth, and Ag 3d region (c) before and (d) after sputtering ∼10 nm depth in the CS–Cu@Ag NP composite. | ||
In order to better understand the CS–Cu@Ag NP composite structure, XPS depth-profile studies were performed by sputtering a small area of the specimen surface (3 keV Ar+ ions for ∼2 min resulting in ∼10 nm depth) and analyzing the freshly exposed surface. The high-resolution XPS data, before and after sputtering, for Ag 3d, are shown in Fig. 3c and d. The binding energy from XPS curves for Ag 3d5/2 and Ag 3d3/2 electrons in Fig. 4c and d, fitted by Gaussian–Lorentzian cross curves, shows two peaks at 368.3 eV and 374.3 eV, which are associated with Ag(0).39,49,52,53 Similarly, Cu 2p signal of the sputtered sample show two peaks at 932.4 eV and 952.2 eV corresponding to the binding energy for Cu 2p3/2 and Cu 2p1/2 (Fig. 3b), similar to those reported for Cu(0).52,53 Relative enhancement of Cu 2p signals as compared to Ag 3d signal for a sputtered surface (refer to ESI, Fig. S6†) indicates the preferential presence of Cu in the core of the sample. Also, absence of separate satellite peaks, corresponding to CuO at ∼10 nm depth-profiled samples of CS–Cu@Ag NP, confirmed that the oxide layer formed only on the exposed surface of the core–shell Cu@Ag NPs in the CS matrix. Combined with TEM images demonstrating average Ag shell thickness of ∼5.8 nm, XPS results of before and after sputtering (∼10 nm) support the core–shell structure of the CS–Cu@Ag NP composite, with the core being Cu(0) and the shell being Ag(0).
3.2 Antibacterial properties of CS–Cu@Ag NP composite
The antibacterial activity of the CS–Cu@Ag NPs was examined on Gram-positive B. cereus and Gram-negative GFP-expressing E. coli by inoculating the bacteria (108 cfu mL−1) in the corresponding media supplemented with various amount of CS–Cu@Ag NPs and allowing to grow overnight at 37 °C. Appropriate control experiments with only acetic acid (at 0.25% used to disperse the CS–Cu@Ag NPs) were also performed. The antibacterial activity of the CS–Cu@Ag NP composite (at MIC and MBC) was checked by measuring the OD at 595 nm at regular intervals and the results on GFP-expressing E. coli are shown in Fig. 4. The bactericidal efficacies of individual constituents of the composite, namely CS–Cu NPs, Ag NPs, CS–Ag NPs and bare Cu@Ag NPs towards the recombinant GFP-expressing E. coli were also compared (refer to ESI, Fig. S7a†). The MIC of the CS–Cu@Ag NP composite against recombinant GFP-expressing E. coli was found to be 63.4 μg mL−1, consisting of 3.03 μg mL−1 of Cu and 1.47 μg mL−1 of Ag. The corresponding MBC (92.99 μg mL−1) contained 4.44 μg mL−1 of Cu and 2.13 μg mL−1 of Ag. The MIC of the CS–Cu@Ag NP composite against B. cereus was found to be 75.46 μg mL−1, consisting of 3.60 μg mL−1 Cu and 1.75 μg mL−1 of Ag (refer to ESI, Fig. S7b†). The corresponding MBC (98.74 μg mL−1) contained 4.71 μg mL−1 of Cu and 2.28 μg mL−1 of Ag.As evident from Fig. 4, control samples of E. coli with 0.02 M acetic acid only in LB medium exhibited no hindrance to growth. Also, the antibacterial activity of Ag NPs (citrate stabilized) at Ag concentrations corresponding to MIC of CS–Cu@Ag NPs was comparable to that of control. Thus, at this low concentration (1.47 μg mL−1) Ag was ineffective in inhibiting bacterial growth. Moreover, for freshly prepared pristine Cu NPs, the MIC value against GFP-expressing E. coli is reported to be 468.2 μg mL−1,2 which is 150 times of the amount of Cu present in the MIC dose of the present CS–Cu@Ag NP composite. Also, as reported in the literature, 100 μg mL−1 of CS–Ag NP or 235.5 μg mL−1 CS–Cu NP composite is required individually to inhibit bacterial growth of GFP-expressing E. coli.2,4 Clearly the CS–Cu@Ag NP composite demonstrated superior bactericidal activity at this low concentrations of Ag and Cu. Enhanced activity of core shell NPs as compared to the parent NPs is well documented in the literature and generally attributed to a combination of strain and ligand effects.54–59 Lattice mismatch at the interface of the core–shell structure leads to strain in Ag shell where the average bond lengths are altered compared to bulk, and alter the electronic states due to heterometallic bonding interactions known as ligand effect. Further, superior bactericidal potency of the CS–Cu@Ag NP composite is also due to the synergistic effect of CS matrix and the core shell structure of the NPs.
Viability of GFP-expressing E. coli, after treatment with CS–Cu@Ag NP (at MIC and MBC) composite for specified duration, was quantified using flow cytometry.1,2 Healthy bacteria with intact cell wall fluoresce only green due to constitutively expressed GFP, while bacteria with compromized cell walls also fluoresce red due to entry and subsequent binding of PI to bacterial DNA. Dead bacteria, in contrast, fluoresce only red color as GFP leaks out of such cells. Lastly, lysed bacteria do not show any kind of fluorescence. The results of the flow-cytometric analysis shown in Fig. 5 and Table 1 revealed that, after 2 h of treatment with the CS–Cu@Ag NP composite, live and viable bacterial population was 69% at MIC. As the duration of treatment increased, viable bacterial population further decreased by ∼10% at MIC dose after 6 h. This was mainly due to an increase in number of compromised bacteria which changed from ∼25% to ∼33%. On the other hand, ca. 99% of bacteria became non-viable after 2 h of treatment at MBC dose. After 6 h of incubation at MBC dose, the only significant change in bacterial population observed was that of an transition from compromised population to dead one (∼17%). On the contrary, the untreated bacteria (control) had less of compromised and dead cells. Overall, the flow-cytometric data suggested that within 2 h of treatment with either MIC or MBC dose of the CS–Cu@Ag NP composite, the bacterial cell wall was damaged irreparably.
| Duration | Viability Stage | Control | MIC | MBC |
|---|---|---|---|---|
| Bacterial population (%) | ||||
| 2 h | Live (LR) | 98.10 | 69.74 | 0.63 |
| Compromised (UR) | 0.18 | 25.46 | 97.59 | |
| Dead (UL) | 0.02 | 3.61 | 1.13 | |
| Lysed (LL) | 1.70 | 1.19 | 0.65 | |
| 4 h | Live (LR) | 98.57 | 65.63 | 0.42 |
| Compromised (UR) | 0.38 | 29.45 | 93.09 | |
| Dead (UL) | 0.01 | 3.71 | 2.85 | |
| Lysed (LL) | 1.04 | 1.21 | 3.64 | |
| 6 h | Live (LR) | 98.12 | 59.31 | 0.23 |
| Compromised (UR) | 0.43 | 33.09 | 77.48 | |
| Dead (UL) | 0.02 | 4.63 | 16.94 | |
| Lysed (LL) | 1.43 | 2.97 | 5.35 | |
3.3 Mechanism of bactericidal action
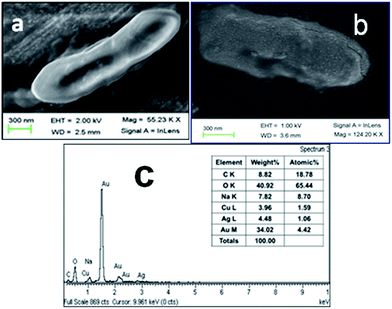
The interaction of the CS–Cu@Ag NP composite and bacterial cells was investigated by FESEM and TEM. It is evident from Fig. 6 that, while untreated (control) bacterial cells were healthy showing regular surface morphology, a treated bacterium had cell wall covered with particles of the CS–Cu@Ag NP composite (Fig. 6b). Corresponding EDX analysis (Fig. 6c) showed the presence of both the elements Cu and Ag, consistent with the core–shell structure of the CS–Cu@Ag NP composite coating the bacteria.The attachment of the composite to the bacteria was also evident in TEM images (Fig. 7) which revealed dark spots of Cu@Ag NP along with CS overlying the surface of a bacterium. The SAED pattern (Fig. 7c) of the particles indicated the presence of crystalline Cu@Ag NPs over the bacterial surface. XRD analysis of bacterial samples (Fig. S8, refer to ESI†), treated with the CS–Cu@Ag NP composite for 12 h, showed the presence of diffraction peaks from the (111), (200), (220) and (311) plane of Ag, indicating that NPs were still present in the treated bacteria.
 | ||
| Fig. 7 (a and b) TEM images of GFP-expressing E. coli treated with MIC of CS–Cu@Ag NP composite for 3 h and (c) corresponding SAED pattern. | ||
The combined results of FESEM, TEM and XRD confirm the interaction of CS–Cu@Ag NP composite with bacterial membrane. CS interacts with bacterial cell wall through electrostatic attractions as CS is polycationic in nature and the bacterial cell envelop is negatively charged.60–64 In the present study, the attachment of CS–Cu@Ag NPs to bacterial cell may cause the permeabilization of the cell wall leading to leakage of intracellular components and eventually cell death. This mechanism is similar to that reported earlier for CS–Ag NP, iodinated CS–Ag NP, and iodinated CS–Cu NP composites.1,2,4 The potential electrostatic interaction between the CS–Cu@Ag NP composite and bacteria was supported by the zeta potential (ζ) measurements which revealed ζ values (at pH 6.3) for CS and CS–Cu@Ag NPs to be +59.57 mV and +23.93 mV, respectively. As previously reported, once attached to the bacterial surface, Ag NPs or CS-based Ag NP composites interact with the sulfur-containing proteins within the bacterial membrane enhancing their permeability and finally leading to leakage of cytoplasmic materials. For a better understanding of this mechanism of bactericidal action in the present study, XPS analysis was carried out on sample of E. coli treated with CS–Cu@Ag NP composite for 24 h. From high resolution XPS analysis, the binding energy for Ag 3d5/2 and Ag 3d3/2 electrons on this sample were found to be 367.43 eV and 373.76 eV, respectively (Fig. 8). The decrease in binding energies of both Ag 3d5/2 and Ag 3d3/2 (corresponding standard binding energies for pure Ag: 368.3 and 374.3 eV, respectively), can be rationalized to be due to the electron transfer between Ag and sulphur.65–68 Thus the XPS results indicated the existence of Ag–S species on the surface of E. coli treated with CS–Cu@Ag NP composite. This result corroborates the fact that the NPs interacted with sulfur-containing proteins present in the bacterial cell membrane making pores in the cell wall. Additionally, the flow-cytometric results and TEM images (Fig. 7) of CS–Cu@Ag NP composite treated bacteria also support our claim of bacterial death as a consequence of damage in cell wall.
 | ||
| Fig. 8 High resolution XPS spectra of Ag 3d region of E. coli sample treated with CS–Cu@Ag NP composite for 24 h. | ||
It may be noted here that the Cu@Ag NPs of the composite, when bound to the cell membrane remained intact, as the particle size distribution of the as prepared sample and the sample attached to the bacterial wall following treatment were comparable (refer to ESI, Fig. S9 and S10†). Thus, neither dissolution nor aggregation of Cu@Ag NPs within the composite had occurred when they were attached to the bacterial cell wall. Thus, in the present study, core–shell Cu@Ag NPs along with CS were mainly responsible for bactericidal activity of CS–Cu@Ag NP composites, rather than the corresponding metal ions.
In order to understand the potential interaction between bacterial DNA and CS–Cu@Ag NP composite, GFP plasmid (pGFP) was isolated from non-treated (control) and CS–Cu@Ag NPs treated recombinant E. coli samples at different time points.45 As evident from the gel electrophoresis results (Fig. S11†), interaction of Cu@Ag NPs and pure plasmid DNA shows retarded movement of pGFP (ESI, Fig. S11a†) depending on the relative amount of the composite present. However, electrophoretic mobility of plasmid isolated from CS–Cu@Ag NP composite-treated bacteria did not reveal any gross variance in migration pattern (refer to ESI, Fig. S11b†). The above agarose gel electrophoresis experiments indicated that CS–Cu@Ag NP possibly had no direct effect on pDNA inside the bacteria. Moreover, the cytotoxicity of CS–Cu@Ag NP composite was tested on normal HEK 293 cells by MTT assay (Fig. 9). It was observed that, while cell viability was almost unaffected at MIC, almost 90% cells were viable even after 48 h at 2× MIC of the composite indicating low-toxicity associated with the composite.
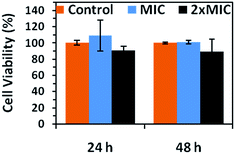 | ||
| Fig. 9 Viability of HEK 293 cells in presence of MIC and 2× MIC of CS–Cu@Ag NP composite after 24 h and 48 h. | ||
4. Conclusions
In summary, we have synthesized CS-stabilized bimetallic core–shell Cu@Ag NPs at ambient conditions by reducing AgNO3 on pre-synthesized CS–Cu NPs. The presence of Ag shell around the core Cu NPs markedly reduced the vulnerability of Cu to aerial oxidation, increasing their stability from 12 h in CS–Cu NPs to more than two weeks in CS–Cu@Ag NPs. The formation of Cu@Ag core–shell structure was confirmed by UV-visible, TEM, HRTEM, XRD and XPS analyses. The NPs in the CS–Cu@Ag NP composite were spherical with an average size of 14 nm. The CS–Cu@Ag NP composite was found to be highly effective in killing Gram-negative E. coli and Gram-positive B. cereus bacteria. The MIC of the CS–Cu@Ag NP composite against E. coli was found to be 63.4 μg mL−1. Bactericidal activity of the composite was found to be superior to those of the individual components due to synergistic effect. Our study suggests that polycationic CS helps to attach the composite into the negatively charged bacterial cell wall. The core–shell Cu@Ag NPs embedded in the composite interact with the proteins of the bacterial membrane leading to the perforation of the cell wall and subsequent leakage of intracellular components. This irreversible damage of the bacterial membrane by the CS–Cu@Ag NP composite results in bacterial death. The present study demonstrates the importance of synthesis and the synergistic antimicrobial activity of CS–Cu@Ag NP composite.Acknowledgements
We thank Department of Science and Technology (DST, SR/S1/PC-30/2008), Department of Biotechnology (DBT, BT/49/NE/TBP/2010) for funds. S.M. thanks CSIR for a fellowship (09/731(0061)/2008-EMR-I). P.S. thanks IIT Guwahati for the Institute Post Doctoral Fellowship. Assistance from Central instruments facility (CIF), Department of Physics for DST-FIST (ref no. SR/FST/PS11-020/2009) X-ray Diffractometer, Centre for the Environment for AAS studies are acknowledged. Mr Subhojit Das, Mr Amaresh Kumar Sahoo, and Ms Upashi Goswami are gratefully acknowledged. Assistance from DST-FIST funded XPS facility at Department of Physics and Meteorology, IIT Kharagpur is also gratefully acknowledged.References
- M. Banerjee, S. Mallick, A. Paul, A. Chattopadhyay and S. S. Ghosh, Langmuir, 2010, 26, 5901 CrossRef CAS PubMed.
- S. Mallick, S. Sharma, M. Banerjee, S. S. Ghosh, A. Chattopadhyay and A. Paul, ACS Appl. Mater. Interfaces, 2012, 4, 1313 CAS.
- S. K. Gogoi, P. Gopinath, A. Paul, A. Ramesh, S. S. Ghosh and A. Chattopadhyay, Langmuir, 2006, 22, 9322 CrossRef CAS PubMed.
- P. Sanpui, A. Murugadoss, P. V. D. Prasad, S. S. Ghosh and A. Chattopadhyay, Int. J. Food Microbiol., 2008, 124, 142 CrossRef CAS PubMed.
- A. K. Sahoo, M. P. Sk, S. S. Ghosh and A. Chattopadhyay, Nanoscale, 2011, 3, 4226–4233 RSC.
- U.S. EPA, National Primary Drinking Water Regulations: Final Rule. Fed, Reg., January 30, 1991, vol. 56(20), pp. 3526–3597 Search PubMed.
- E. Gaggelli, H. Kozlowski, D. Valensin and G. Valensin, Chem. Rev., 2006, 106, 1995 CrossRef CAS PubMed.
- M. A. Cater, S. La Fontaine, K. Shield, Y. Deal and J. F. B. Mercer, Gastroenterology, 2006, 130, 493 CrossRef CAS PubMed.
- G. Ren, D. Hu, E. W. C. Cheng, M. A. Vargas-Reus, P. Reip and R. P. Allaker, Int. J. Antimicrob. Agents, 2009, 33, 587 CrossRef CAS PubMed.
- R. H. Morriss and L. F. Collins, J. Chem. Phys., 1964, 41, 3357 CrossRef CAS PubMed.
- N. Toshima and T. Yonezawa, New J. Chem., 1998, 22, 1179 RSC.
- M. Grouchko, A. Kamyshny and S. Magdassi, J. Mater. Chem., 2009, 19, 3057 RSC.
- M. Cazayous, C. Langlois, T. Oikawa, C. Ricolleau and A. Sacuto, Phys. Rev. B: Condens. Matter Mater. Phys., 2006, 73, 113402 CrossRef.
- M. Tsuji, S. Hikino, R. Tanabe, M. Matsunaga and Y. Sano, CrystEngComm, 2010, 12, 3900 RSC.
- J. Zhang, Y. Tang, K. Lee and M. Ouyang, Science, 2010, 327, 1634 CrossRef CAS PubMed.
- B. Rodriguez-Gonzalez, A. Burrows, M. Watanabe, C. J. Kiely and L. M. Liz Marzan, J. Mater. Chem., 2005, 15, 1755 RSC.
- Y. Ma, W. Li, E. C. Cho, Z. Li, T. Yu, J. Zeng, Z. Xie and Y. Xia, ACS Nano, 2010, 4, 6725 CrossRef CAS PubMed.
- I. Srnova-Sloufova, F. Lednicky, A. Gemperle and J. Gemperlova, Langmuir, 2000, 16, 9928 CrossRef CAS.
- A. Henglein, J. Phys. Chem. B, 2000, 104, 6683 CrossRef CAS.
- R. Fernando, J. Jellinek and R. L. Johnston, Chem. Rev., 2008, 108, 845 CrossRef PubMed.
- N. L. Netzer, C. Qiu, Y. Zhang, C. Lin, L. Zhang, H. Fong and C. Jiang, Chem. Commun., 2011, 47, 9606 RSC.
- C. Langlois, D. Alloyeau, Y. Le Bouar, A. Loiseau, T. Oikawa, C. Mottet and C. Ricolleau, Faraday Discuss., 2008, 138, 375 RSC.
- V. R. Mancier, C. l. Rousse-Bertrand, J. Dille, J. Michel and P. Fricoteaux, Ultrason. Sonochem., 2010, 17, 690 CrossRef CAS PubMed.
- M. Tsuji, S. Hikino, Y. Sano and M. Horigome, Chem. Lett., 2009, 38, 518 CrossRef CAS.
- T. S. Anderson, R. H. Magruder Iii, J. E. Wittig, D. L. Kinser and R. A. Zuhr, Nucl. Instrum. Methods Phys. Res., Sect. B, 2000, 171, 401 CrossRef CAS.
- D. Manikandan, S. Mohan, P. Magudapathy and K. G. M. Nair, Nucl. Instrum. Methods Phys. Res., Sect. B, 2002, 198, 73 CrossRef CAS.
- D. Manikandan, S. Mohan and K. G. M. Nair, Phys. B, 2003, 337, 64 CrossRef CAS.
- X. Xu, X. Luo, H. Zhuang, W. Li and B. Zhang, Mater. Lett., 2003, 57, 3987 CrossRef CAS.
- D. S. Jung, H. M. Lee, Y. C. Kang and S. B. Park, J. Colloid Interface Sci., 2011, 364, 574 CrossRef CAS PubMed.
- J. Zhao, D. Zhang and X. Song, Appl. Surf. Sci., 2012, 258, 7430 CrossRef CAS PubMed.
- N. Toshima, M. Harada, Y. Yamazaki and K. Asakura, J. Phys. Chem., 1992, 96, 9927 CrossRef CAS.
- N. Toshima, Pure Appl. Chem., 2000, 72, 317 CrossRef CAS.
- Y. Mizukoshi, T. Fujimoto, Y. Nagata, R. Oshima and Y. Maeda, J. Phys. Chem. B, 2000, 104, 6028 CrossRef CAS.
- S. Rana, J. Rawat and R. D. K. Misra, Acta Biomater., 2005, 1, 691 CrossRef CAS PubMed.
- B. Chudasama, A. Vala, N. Andhariya, R. V. Upadhyay and R. V. Mehta, Nano Res., 2010, 2, 955 CrossRef.
- Y. H. Kim, D. K. Lee, H. G. Cha, C. W. Kim and Y. S. Kang, J. Phys. Chem. C, 2007, 111, 3629 CAS.
- P. Gong, H. M. Li, X. X. He, K. M. Wang, J. B. Hu, W. H. Tan, S. C. Zhang and X. H. Yang, Nanotechnology, 2007, 18, 28560 Search PubMed.
- M. Banerjee, S. Sharma, A. Chattopadhyay and S. S. Ghosh, Nanoscale, 2011, 3, 5120 RSC.
- M. Taner, N. Sayar, I. G. Yulug and S. Suzer, J. Mater. Chem., 2011, 21, 13150 RSC.
- M. Valodkar, S. Modi, A. Pal and S. Thakore, Mater. Res. Bull., 2011, 46, 384 CrossRef CAS PubMed.
- J. T. Trevors and C. M. Cotter, J. Ind. Microbiol., 1990, 6, 77 CrossRef CAS.
- Q. L. Feng, J. Wu, G. Q. Chen, F. Z. Cui, T. N. Kim and J. O. Kim, J. Biomed. Mater. Res., 2000, 52, 662 CrossRef CAS.
- R. L. Williams, P. J. Doherty, D. G. Vince, G. J. Grashoff and D. F. Williams, Crit. Rev. Biocompat., 1989, 5, 221 CAS.
- J. Lehtinen, J. Nuutila and E.-M. Lilius, Cytometry, Part A, 2004, 60, 165 CrossRef PubMed.
- H. C. Birnboim and J. Doly, Nucleic Acids Res., 1979, 7, 1513 CrossRef CAS PubMed.
- J. Sambrook and D. W. Russell, Molecular Cloning-A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York, 3rd edn, 2001, vol. 1, pp. 1.31–1.38 Search PubMed.
- D. Mott, J. Galkowski, L. Wang, J. Luo and C.-J. Zhong, Langmuir, 2007, 23, 5740 CrossRef CAS PubMed.
- L. Lu, W. Zhang, D. Wang, X. Xu, J. Miao and Y. Jiang, Mater. Lett., 2010, 64, 1732 CrossRef CAS PubMed.
- J. Zhao, D. Zhang and J. Zhao, J. Solid State Chem., 2011, 184, 2339 CrossRef CAS PubMed.
- J. Torres, E. Valles and E. Gomez, J. Nanopart. Res., 2010, 12, 2189 CrossRef.
- Y. H. Kim, D. K. Lee, H. G. Cha, C. W. Kim, Y. C. Kang and Y. S. Kang, J. Phys. Chem. B, 2006, 110, 24923 CrossRef CAS PubMed.
- M. P. Seah, I. S. Gilmore and G. Beamson, Surf. Interface Anal., 1998, 26, 642 CrossRef CAS.
- J. F. Moulder and J. Chastain, Handbook of X Ray Photoelectron Spectroscopy: A Reference Book of Standard Spectra for Identification and Interpretation of XPS Data, Perkin-Elmer, 1995 Search PubMed.
- R. Burch, Acc. Chem. Res., 1982, 15, 24 CrossRef CAS.
- M. Mavrikakis, B. Hammer and J. K. Norskov, Phys. Rev. Lett., 1998, 81, 2819 CrossRef.
- J. R. Kitchin, J. K. Norskov, M. A. Barteau and J. G. Chen, Phys. Rev. Lett., 2004, 93, 156801 CrossRef CAS.
- J. R. Kitchin, J. K. Norskov, M. A. Barteau and J. G. Chen, J. Chem. Phys., 2004, 120, 10240 CrossRef CAS PubMed.
- J. X. Wang, H. Inada, L. Wu, Y. Zhu, Y. Choi, P. Liu, W.-P. Zhou and R. R. Adzic, J. Am. Chem. Soc., 2009, 131, 17298 CrossRef CAS PubMed.
- Y. Xing, Y. Cai, M. B. Vukmirovic, W.-P. Zhou, H. Karan, J. X. Wang and R. R. Adzic, J. Phys. Chem. Lett., 2010, 1, 3238 CrossRef CAS.
- N. R. Sudarshan, D. G. Hoover and D. Knorr, Food Biotechnol., 1992, 6, 257 CrossRef CAS.
- I. M. Helander, E.-L. Nurmiaho-Lassila, R. Ahvenainen, J. Rhoades and S. Roller, Int. J. Food Microbiol., 2001, 71, 235 CrossRef CAS.
- E. I. Rabea, M. E.-T. Badawy, C. V. Stevens, G. Smagghe and W. Steurbaut, Biomacromolecules, 2003, 4, 1457 CrossRef CAS PubMed.
- J. Li and L. A. McLandsborough, Int. J. Food Microbiol., 2001, 53, 185 CrossRef.
- W. W. Wilson, M. M. Wade, S. C. Holman and F. R. Champlin, J. Microbiol. Methods, 2001, 43, 153 CrossRef CAS.
- F. Bensebaa, Z. Yu, Y. Deslandes, E. Kruus and T. H. Ellis, Surf. Sci., 1998, 405, L472–L476 CrossRef CAS.
- M. J. Esplandiu and P. L. M. Noeske, Appl. Surf. Sci., 2002, 199, 166 CrossRef CAS.
- R. Chen, N. T. Nuhfer, L. Moussa, H. R. Morris and P. M. Whitmore, Nanotechnology, 2008, 19, 455604 CrossRef PubMed.
- H. Shengtai, Y. Jiannian, X. Sishen, G. Hongjun and P. Shijin, J. Phys. D: Appl. Phys., 2001, 34, 3425 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Additional SEM, TEM, XRD, FTIR, XPS, antibacterial and agarose gel electrophoresis results are available. See DOI: 10.1039/c4ra12770f |
| This journal is © The Royal Society of Chemistry 2015 |