Dispersive and FT-Raman spectroscopic methods in food analysis†
Ismail Hakki Boyaci*ab,
Havva Tümay Temiza,
Hüseyin Efe Genişa,
Esra Acar Soykutb,
Nazife Nur Yazgana,
Burcu Güvena,
Reyhan Selin Uysala,
Akif Göktuğ Bozkurta,
Kerem İlaslana,
Ozlem Toruna and
Fahriye Ceyda Dudak Şekera
aDepartment of Food Engineering, Faculty of Engineering, Hacettepe University, Beytepe, 06800 Ankara, Turkey. E-mail: ihb@hacettepe.edu.tr; Fax: +90 312 299 21 23; Tel: +90 312 297 61 46
bFood Research Center, Hacettepe University, Beytepe, 06800 Ankara, Turkey
First published on 22nd June 2015
Abstract
Raman spectroscopy is a powerful technique for molecular analysis of food samples. A fingerprint spectrum can be obtained for a target molecule by using Raman technology, as specific signals are obtained for chemical bonds in the target. In this way, food components, additives, processes and changes during shelf life, adulterations and numerous contaminants such as microorganisms, chemicals and toxins, can also be determined with or without the help of chemometric methods. The studies included in this review show that Raman spectroscopy has great potential for food analyses. In this review, we aim to bring together Raman studies on components, contaminants, raw materials, and adulterations of various food, and attempt to prepare a database of Raman bands obtained from food samples.
Introduction
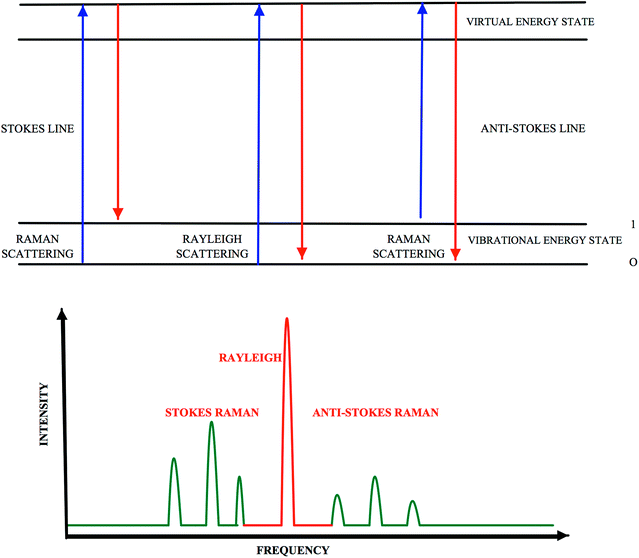
Customers have recently become more concerned about being informed on food quality and, thus the need for food quality determination methods has increased. Therefore, all steps in food production such as composition or quality of the products, their origin and how they have been handled, processed and stored have gained importance. Various types of methods, including microbial methods, sensory analysis, biochemical and physicochemical methods are used in food analysis. Chromatographic methods such as high-performance liquid chromatography (HPLC) and gas chromatography (GC) have become very popular for separation and identification of food components due to their high reproducibility and low detection limits.1 Deoxyribonucleic acid (DNA) based methods, such as polymerase chain reaction (PCR) techniques, and immunological based methods, such as enzyme-linked immunosorbent Assay, are also used for detection of specific targets in food samples. Although these are common methods being used, they cannot meet the demand for in situ, rapid and multiple analysis. Spectrophotometric methods, on the other hand, has a great advantage over them as it successfully meets those requirements. Spectroscopic methods which are used to determine different properties of food components is a rapid and easy method.2 The spectroscopic methods used for food analysis include ultra violet-visible spectroscopy (UV-Vis spectroscopy), fluorescence spectroscopy, Raman spectroscopy and infrared spectroscopy (IR), circular dichroism (CD), X-ray spectroscopy, nuclear magnetic resonance, electron spin resonance, dielectric spectroscopy, and photoacoustic Spectroscopy.Raman spectroscopy detects chemical and organic molecule types and their physical structures by making use of bonds.2 Photons are scattered when an intense monochromatic light source – especially a laser beam – irradiates a sample, and as a result of this application, the largest fraction of the scattered light is found to have the same wavelength as the laser light. This elastic scattering is known as Rayleigh scattering. Far fewer inelastic collisions occur between the sample and the incident photons. As a result, the wavelengths of scattered photons change, which is referred to as Raman scattering (Fig. 1). If a molecule gains energy during Raman scattering, the scattered photons will shift to longer wavelength and give Stokes lines; however, if it loses energy, they may shift to shorter wavelengths and give anti-Stokes lines, as seen in Fig. 1.3 The quantity of normal vibrations, masses and geometric arrangements of the atoms in the molecule, as well as the strengths of the chemical bonds between the atoms, form the vibrational spectrum or fingerprint of each molecule. The term fingerprint is justified by the fact that no two samples or compounds have the same spectrum in terms of frequency and intensity of peaks and shoulders.4 Molecules have a large number of vibrational states, but not all molecules are able to have a Raman spectrum. Being Raman active is the basic requirement for a molecule to have a Raman spectrum. Raman spectroscopy can examine molecular vibrations that cause change in their polarizability.5 The symmetry of a molecule is also an important requirement for Raman spectra, since symmetric stretches are more intense in Raman spectra than in Infrared spectra. Functional groups such as C–X (X = F, Cl, Br or I), C–NO2, C–S, S–S, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C
C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C
C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, etc., show greater polarizability changes and give strong Raman signals.5
N, etc., show greater polarizability changes and give strong Raman signals.5
A Raman spectrum consists of scattering intensity (photons per second) plotted vs. wavelength (nanometres) or Raman shift (in reciprocal centimetres). Each band in a Raman spectrum corresponds to a Raman shift occurring due to the incident light energy.6 These bands correspond to specific bands of chemical bonds and/or functional groups of the molecule. By using these specific bands, the fingerprint of a molecule can be obtained with Raman spectroscopy. In addition, quantitative analyses can also be performed by using Raman spectroscopy since the intensity of the band is linearly proportional to the concentration of an analyte.3
Raman spectroscopy has great potential for biochemical analysis. Major advantages of this technique are its ability to provide information about concentration, structure, and interaction of biochemical molecules within intact cells and tissues non-destructively. In addition, it doesn't require homogenization, extraction, the use of dyes or any other labelling agent,7 or any pre-treatment of samples; it only requires small portions of samples. Raman spectroscopy can analyse samples in both liquid and solid phases at ambient temperature and pressure.8 Besides, Raman spectroscopy is a potential tool for the assessment of food quality systems during handling, processing and storage.9
In this review, we will discuss applications of Raman spectroscopy in food analysis (Fig. 2). Conventional analysis methods will be taken into consideration under the headings related to each basic food sample. These headings are listed as Raman Spectroscopy for detection of food components, microorganisms and chemicals in food, food additives, raw materials and food adulterations; and relevant studies are presented under each of them. In addition, vitamins and minerals will be also considered although they found little interest in food analysis by Raman despite of their major role in nutrition and health. There are several reviews in literature about usage of Raman Spectroscopy for food analysis focusing on specific food groups or specific components found in foods.10–13 This review will provide a general insight into the Raman analysis of major components and contaminants found in foods including adulterations. Besides, Raman bands of the important compounds found in food matrix will be presented in a database, which will be helpful for the user of the Raman spectroscopy to analyze new spectra obtained from food samples. Although surface-enhanced Raman spectroscopy (SERS) is a commonly used method for improving the sensitivity of Raman spectroscopy, studies on SERS haven't been included in this review since it requires a specific surface and a Raman active molecule in order to provide enhancement and to obtain characteristic Raman signals.
Raman spectroscopy for the detection of food components
Composition of the food samples has great importance due to its substantial effect on quality, nutritional and economic value, and its contribution to the properties of final product. Environmental factors and applied processing factors could have both positive and negative effects on food components. Therefore, monitoring these changes in every step of food production process is of great importance. There are many ways to determine these changes in food components, and for this purpose, there has been an increasing interest in the use of Raman spectroscopy over the past few decades. In this section, an overview is given on the use of Raman spectroscopy in quantitative and qualitative analysis of food components. Specific Raman bands related to the articles investigated in this section were given in Tables 1S–4S† for proteins, carbohydrates, lipids and vitamins, respectively.Proteins
Raman spectroscopy enables researchers to obtain detailed information on structural properties of proteins. Studies on food proteins have been conducted for several decades, and they still maintain their importance since proteins are one of the major components of foods and have important effects on properties of food. Proteins are large polypeptides consisting of hundreds of amino acids; thus, a complex set of overlapping bands forms their Raman spectra. Additionally, strong Raman scattering of aromatic amino acids and polypeptide chains also contribute to the existence of characteristic bands observed in the Raman spectra.14Raman spectroscopy has been used in food protein studies on various topics, but in particular, investigation of the secondary structure of proteins has been the focus of them. In these studies, amide bands-especially the amide I (1645–1685 cm−1) and amide III (1200–1350 cm−1) bands have proven to be the most useful bands to obtain data on the secondary structures of proteins, which are composed of α-helices, β-sheets, turns and random coil structures. Characteristic Raman bands of various amino acid residues have also been used to acquire information about the microenvironment and conformational changes of the proteins. For instance, phenylalanine residue has been generally used as an internal standard in most of the Raman spectroscopic studies15–17 since it is reported to be insensitive to conformation or microenvironment.18 Chi et al. used the advantages of UV-resonance Raman spectroscopy (UV-RR spectroscopy), such as its ability to selectively examine the secondary structures of dilute protein and peptide solutions.19 Chemometric methods were employed to determine the average amide band resonance Raman spectra of the α-helix, β-sheet, and unordered secondary structures of a number of proteins. Similarly, Huang, Balakrishnan, and Spiro used deep-UV-RR spectroscopy to investigate the secondary structures of proteins.20 Resonance-enhanced amide bands and aromatic side chain bands of proteins with varied secondary structure contents were analysed with least-squares fitting method to establish quantitative signatures of secondary structure.
Several reactions occurring in the food matrix due to application of different processing techniques could be monitored by Raman spectroscopy. Nonaka, Li-Chan, and Nakai studied thermal-induced gelation process of whey proteins. Different time and temperature parameters were applied to α-lactoglobulin and β-lactoglobulin proteins, and changes in their Raman spectra were followed.21 In another study, Raman spectroscopy was used to investigate the interaction of lysozyme, α-lactoglobulin and β-lactoglobulin before and after the completion of gelation process.22 Rheological changes and the interactions of egg albumen and whey protein during the gelation process were tracked with FT-Raman spectroscopy using phenylalanine (1004 cm−1) as an internal standard. Differences in the gel structures were evaluated by monitoring CH (1350 cm−1) and CH2 (1450 cm−1) bending vibrations. As a result, an increment was observed for the Raman intensity of β-sheet structures in the amide III region, while the intensities decreased for helical structures.17 Sánchez-González et al. used FT-Raman to examine the structural changes in proteins and water during gelation of fish surimi. Amide bands and amino acid residue bands were monitored to detect the changes.23 The changes in chemical structures occurring due to heat treated gelatinization of myofibrillar proteins contribute to the quality of meat products and to improve their production techniques. Hence, conformational changes in peptide structures of meat, which is highly rich in protein, were tracked by using Raman spectroscopy. It was found that owing to the impact of heat, changes in amide I (1600–1700 cm−1) and amide III (1200–1300 cm−1) regions reduces the α-helices content in protein structure, while it increases β-sheets, β-turns and random coil content.24 In another study, whether endpoint temperature (EPT) was reached for cooked meat and meat products was checked by using Raman Spectroscopy with chemometric analysis. It was found that secondary structure of meat proteins were changed with heat treatment as in the previous study.25 Lim et al., studied gelation of phenol extracted protein fractions from non-acclimated (NA) and cold-acclimated (CA) winter rye leaf tissue after repeated freeze–thaw treatments. Changes in the protein secondary structure caused by the freeze–thaw cycles were monitored by using Raman microscopy. Gelling and non-gelling components as well as protein extracts were individually analyzed with Raman measurements. Similarity between NA and CA samples was explained with their similar structural conformations, which were stabilized by similar protein–protein interactions, while dissimilarities were mainly assigned to the covalent bonds altered by freeze–thaw treatments.26 Lactate dehydrogenase was chosen as a model protein to evaluate the potential of Raman spectroscopy for discriminating native like and non-native states of protein in freeze-dried formulations. PCA and PLS–LDA methods were applied to collected Raman data for discrimination studies. Different prediction models were developed using different spectral regions and their combinations. C–N stretch, NH3 deformation, amide III, and mainly C–Hn non-stretching with possible participations of C–N and C–C stretching were considered to have the maximum contribution to the success of discrimination.27
Deamidation of proteins is another process which can be easily followed by using Raman spectroscopy. Wong et al. used soy and whey protein isolates and spray-dried egg white powder to analyze the extent of deamidation in food proteins. Conformational changes of the protein structures were examined using Raman spectroscopy. Characteristic band, which was assigned to the stretching of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond of glutamate or aspartate, was used as the marker band for deamidation.28 In another work, the enzymatic hydrolysis of wheat gluten substrates, which were acid-deamidated by using three different acids were determined with Raman spectroscopy. The same degree of deamidation with the same heat treatment conditions was applied to all substrates. Raman spectroscopic analyses of microenvironments belonging to Cys, Trp, Tyr and His amino acids showed a positive relation with their susceptibilities to enzymatic hydrolysis.16
O bond of glutamate or aspartate, was used as the marker band for deamidation.28 In another work, the enzymatic hydrolysis of wheat gluten substrates, which were acid-deamidated by using three different acids were determined with Raman spectroscopy. The same degree of deamidation with the same heat treatment conditions was applied to all substrates. Raman spectroscopic analyses of microenvironments belonging to Cys, Trp, Tyr and His amino acids showed a positive relation with their susceptibilities to enzymatic hydrolysis.16
Kang et al. used Raman spectroscopy to evaluate the effects of salt content and also chopping and beating processes on the structural components of pork frankfurters. In the case of increased salt content, a decrease was observed for C–H stretching, CH2 and CH3 bending vibrations, while no changes were witnessed for secondary structures, namely tryptophan, tyrosine residues, β-sheet and α-helix. Beating process was resulted with an increase in β-sheet, a decrease in α-helix content and a decrease in C–H stretching, CH2 and CH3 bending vibrations. The adequacy of Raman spectroscopy as the experimental technique to follow the changes (appearing and disappearing of compounds) in composition during the beating process was demonstrated in this study.29
The interactions of proteins with other food components are another topic of interest commonly dealt with in using studies Raman spectroscopy. Shao et al., for instance, used Raman Spectroscopy to examine the emulsion created by adding lipid to meat. They followed the changes occurring in protein structures due to addition of different lipids to meat and heat treatment. They found that there wasn't a considerable change in the secondary structures of proteins in emulsion spectra obtained by mixing three different kinds of lipids without any heat treatment. With the application of heat treatment, however, it was seen that changes occurred in the bands (1153 cm−1) demonstrating amide I (1654 cm−1), amide II (1517 cm−1), amide III (1300 cm−1) and C–N stretching vibration, and that formation of β-sheet increased. In this way, the researchers put forward the idea that with Raman spectroscopy, protein–lipid–water interactions could be examined and information could be obtained directly.30 Meng et al., however, examined the protein–lipid interaction with Raman microscopy by using bovine serum albumin–oil. Different spectra were obtained by extracting the spectrum of mineral or corn oil from that of the bovine serum albumin (BSA)–oil interface in order to determine the contributions of different functional groups to the protein–lipid interactions.31 In the study of Sivam et al., Raman and FT-IR spectroscopy were used as complementary methods to explore the conformational changes in wheat proteins and polysaccharides due to their interactions with fruit polyphenols and pectin. Amide bands, in particular, were examined to comprehend the interactions between additives and gluten proteins by following changes in the secondary structures of the proteins.32 In the study of Ferrer et al., gluten protein was chemically modified with an emulsifier, namely sodium stearoyl lactylate (SSL), and the effect of the modification on the secondary and tertiary structures of this protein was analysed with FT-Raman spectroscopy. A significant increase was observed in the intensity of amide I band, which is attributed to a more ordered structure. Conformational variations of disulfide bounds and the variations of the intensity rate of the tyrosine doublet bands, tryptophan and C–H stretching band were also explained by formation of a more ordered structure.15 In the subsequent study of the same group, Gomez et al. reported a more detailed research investigating the conformational changes in the presence of another emulsifier, namely diacetyl tartaric acid esters of monoglycerides: DATEM. A comparison was also made on the differences in the gluten structures occurring in the presence of DATEM and SSL, respectively. Differences on Raman spectra were mainly assigned to the distinct chemical structures of these emulsifiers which specify their type of interactions with gluten proteins.33 Perisic et al. used the combinations of vibrational spectroscopic techniques, namely NIR spectroscopy and FT-IR, NIR and Raman microspectroscopy to study the effects of different salts on the hydration properties of structural proteins. Interactions between salt cations and aromatic amino acid residues were investigated, and their effect on the final structure of proteins was emphasized. Effect of salt concentration on the protein structures were mainly monitored with tyrosine bands which were in a positive correlation with hydrogenated N–H groups.34
Carbohydrates
Structural characterization of carbohydrates is of great importance since they form the widest class of organic compounds. Mono-, di-, oligo- and polysaccharides show characteristic Raman bands by which carbohydrates can be easily determined and quantified. Presence of large number of atoms in the repeat unit and absence of a well-defined entity increased the importance of accurate assignment of vibrational modes in the structural analysis of carbohydrates by using Raman spectroscopy.35 For instance, the amylose contents of corn and cassava starch samples were quantified using FT-Raman spectroscopy coupled with PCA and PLS regression methods. Characteristic band at 480 cm−1 was assigned to the ring vibration of starches and used to identify the presence of starch and to distinguish between corn and cassava starch samples.36 Delfino et al. quantified the glucose contents in commercial sports drinks by using micro-Raman and interval partial least square regression (iPLS). Fingerprint bands of glucose, fructose and sucrose were obtained in the spectral region between 600 and1600 cm−1.37 In a recent study reported by our research group, Ilaslan et al. used Raman spectroscopy to quantify glucose, fructose and sucrose contents of commercial soft drinks. Additives in the content of soft drinks were also characterized by their bands, which were assigned to the presence of aroma compounds and citric acid in the composition. The calibration curves were obtained for each component by applying PLS regression on Raman data, and validation studies were carried out using HPLC.38Raman spectroscopy was also used for the detailed investigation of structural components of the food samples. In the study of Roman et al., the components of wild carrot root, such as starch, pectin, cellulose, lignin, and even bioactive polyacetylenes were measured in situ and without any sample preparation. They also showed tissue-specific accumulations of the components using a Raman mapping technique.39 In a similar study, components of wheat and barley grain were investigated using Raman microscopy. The Raman spectra of the most important substances such as proteins, carbohydrates (arabinoxylan, β-glucan and starch) and phytic acid were included in the compositions of barley, and wheat cells were measured. Wheat proteins were monitored by using the characteristic bands of gluten located around 1449 and 1659 cm−1, which was attributed to the CH2 bending mode of amino acids and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching mode of amides, respectively. Polysaccharides, namely arabinoxylan, β-glucan and starch gave similar Raman spectra. Distinct bands at 1095 and 1120 cm−1 assigned to the COC stretching vibrations of glycosidic bonds showed the most evident similarity between these spectra. Starch was separated from the other polysaccharides by its characteristic bands at 480 and 901 cm−1 (skeletal vibrations of the glucopyranose ring), and phytic acid gave relatively weak Raman scattering with a characteristic band at 3420 cm−1 (stretching of OH group). Raman imaging was also performed to analyze the distribution of these components in the cereal grain structure.40
O stretching mode of amides, respectively. Polysaccharides, namely arabinoxylan, β-glucan and starch gave similar Raman spectra. Distinct bands at 1095 and 1120 cm−1 assigned to the COC stretching vibrations of glycosidic bonds showed the most evident similarity between these spectra. Starch was separated from the other polysaccharides by its characteristic bands at 480 and 901 cm−1 (skeletal vibrations of the glucopyranose ring), and phytic acid gave relatively weak Raman scattering with a characteristic band at 3420 cm−1 (stretching of OH group). Raman imaging was also performed to analyze the distribution of these components in the cereal grain structure.40
Application of Raman spectroscopy makes it possible to determine the sources of carbohydrates and extract minor differences between very similar structures. Scudiero and Morris used Raman spectroscopy to identify the differences between soft and hard wheat flour samples. Relative intensity ratios of the bands between 400–600 cm−1 and 1020–1650 cm−1 corresponding to the arabino-to-xylan substitution and phenolic acid contents were used to differentiate the samples.41 Wellner et al. compared the composition and physical structure of starch granules found in wild type and mutant maize kernels by using a Raman imaging technique.42 Similar characteristic bands of carbohydrates and protein structures, specifically amide I bands were observed for both wild type and mutant samples. However, differences originating from the variations in the ratio of branched residues to linear residues were monitored by following the characteristic band at 942 cm−1, which was reported as being sensitive to the level of branching in starch polysaccharide. Another characteristic band at 865 cm−1 was used to monitor the crystalline structure of starch granules.43 Compositional and structural properties of β-glucan in barley and oat samples were investigated with FT-Raman spectroscopy. PCA and PLS regression were used for multivariate data analysis of collected Raman data, especially in the spectral region between 800 and 1800 cm−1. PLS regression prediction models successfully determining the β-glucan and starch contents of the samples were created. Clusters of cellulose, curdlan and cellulose–curdlan blends were located in the PCA score plot depending on the variation in their β-glucan structure.44
The effects of food processing on carbohydrates, one of which is starch modification, have also been analysed with Raman spectroscopy. Chong et al. determined the degree of maleate substitution in maleinated starches depending on the emergence of new bands, which were likely due to nominal C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch, C
O stretch, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch, and O–H stretch vibrational modes.45 Another modification of starch was carried out with octenyl succinate. This modification treatment was monitored using Raman microscopy integrated with AFM.46 The use of Raman spectroscopy for quality control of modified starches in the food industry was demonstrated by Dupuy and Laureyns. They identified the modified starches according to their origin and type of modification. Although an overall similarity was observed for different starch samples, disappearance of the doublet at 600 cm−1 was observed for pregelatinized samples. Similarly, waxy samples were monitored by their characteristic bands at 480, 870, 950 and 1468 cm−1, which were assigned to skeletal mode, the CH and CH2 deformation, the skeletal mode involving α(1–4) linkage and the CH2 deformation, respectively. They also compared the efficiency of the chemometric methods PCA and PLS in order to group the samples according to the applied modification type and draw the conclusion that PLS is more effective than PCA.47 Passauer et al. studied the degrees of substitution (DS) of starch phosphates by using the characteristic band of C–O–P stretching vibration at about 975 cm−1.48 Volkert et al. also determined the DS values of different substituted starch acetates by using a combination of FT-Raman spectroscopy and chemometrics. They found the best congruence between determined and calculated DS values by calculating the first derivatives of the Raman spectra.49 In another study, DS values for cationic quaternary ammonium starches were determined using the characteristic band of trimethyl ammonium substituent about 761 cm−1.50 Similarly, the DS values for carboxymethylated non-starch polysaccharides including cellulose, guar gum, locust bean gum and xanthan gum were determined with Raman spectroscopy and a colorimetric method. The characteristic band at 1607 cm−1 was chosen to be the marker of carboxymethylation which originates from C
C stretch, and O–H stretch vibrational modes.45 Another modification of starch was carried out with octenyl succinate. This modification treatment was monitored using Raman microscopy integrated with AFM.46 The use of Raman spectroscopy for quality control of modified starches in the food industry was demonstrated by Dupuy and Laureyns. They identified the modified starches according to their origin and type of modification. Although an overall similarity was observed for different starch samples, disappearance of the doublet at 600 cm−1 was observed for pregelatinized samples. Similarly, waxy samples were monitored by their characteristic bands at 480, 870, 950 and 1468 cm−1, which were assigned to skeletal mode, the CH and CH2 deformation, the skeletal mode involving α(1–4) linkage and the CH2 deformation, respectively. They also compared the efficiency of the chemometric methods PCA and PLS in order to group the samples according to the applied modification type and draw the conclusion that PLS is more effective than PCA.47 Passauer et al. studied the degrees of substitution (DS) of starch phosphates by using the characteristic band of C–O–P stretching vibration at about 975 cm−1.48 Volkert et al. also determined the DS values of different substituted starch acetates by using a combination of FT-Raman spectroscopy and chemometrics. They found the best congruence between determined and calculated DS values by calculating the first derivatives of the Raman spectra.49 In another study, DS values for cationic quaternary ammonium starches were determined using the characteristic band of trimethyl ammonium substituent about 761 cm−1.50 Similarly, the DS values for carboxymethylated non-starch polysaccharides including cellulose, guar gum, locust bean gum and xanthan gum were determined with Raman spectroscopy and a colorimetric method. The characteristic band at 1607 cm−1 was chosen to be the marker of carboxymethylation which originates from C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O carbonyl stretching vibration. The intensity ratios of the marker bands to that of an internal standard band corresponding to the skeletal configuration and linkages (850–950 cm−1) were used to establish a calibration between spectroscopic and colorimetric DS values.51
O carbonyl stretching vibration. The intensity ratios of the marker bands to that of an internal standard band corresponding to the skeletal configuration and linkages (850–950 cm−1) were used to establish a calibration between spectroscopic and colorimetric DS values.51
In another study, the technical starch hydrolysis process was monitored with FT-Raman.52 Gelatinization, liquefaction, saccharification and retrogradation processes were evaluated within the context of the relevant study. The intensity of the bands at 1633 and 3213 cm−1 increased during the gelatinization process, while the others decreased. Liquefaction was characterized by the disappearance of the bands at 735 cm−1 and 480 cm−1. Changes in saccharification at bands in 910–935 cm−1 region and at 1127 cm−1 were also monitored.53,54
Mutungi et al. demonstrated the utility of the FT-Raman method for rapidly determining starch crystallinity, which is important for food production and storage. In this method, a band assigned to symmetric C(1)–O–C(5) stretching of the α-D-glucose ring was used as an internal standard to normalize the spectra. As a result, a strong linear correlation was found between crystallinity and the integrated area of the skeletal mode Raman band.55 Similarly, in the study of Islam and Langrish, Raman spectroscopy was used to investigate the formation of lactose anomers and degree of lactose crystallization during spray drying. The characteristic bands in the Raman spectra indicated the presence of different lactose anomeric and crystalline forms. These bands at 1100 and 350 cm−1 were assigned to the stretching and bending vibrations of the C–O–C grouping of α- and β-lactose structures. Spectral region between 1200 and 1500 cm−1 was used to characterize the presence of an amorphous polymorph in the lactose samples.56 In another study on crystallinity, Raman spectroscopy was used to determine the viscoelastical properties of modified cellulose, which is a significant substance in food industry. Akinosho et al. investigated the effect of methyl and hydroxypropyl groups on gel properties of hydroxypropyl methylcellulose (HPMC). Subsequent to the analysis of collected Raman data, usability of the hydroxypropyl groups as an indicator of the crystalline structure of HPMC was reported. Crystallinity was also monitored by following the significant broadening, which is generally assigned to the decrease in crystallinity in the spectral region between 1540–1660 cm−1.57
Kizil and Irudayaraj evaluated the potential of Raman spectroscopy to follow the chemical changes induced by the application of gamma-irradiation to food samples. Fructose and honey samples were analysed using FT-Raman spectroscopy and canonical discriminant analysis was applied to the collected data. Monitoring the CH stretch region between 2800 and 3000 cm−1, they classified honey samples according to applied irradiation dose. Using spectral regions below 700 cm−1 and between 800 and 1500 cm−1, changes in the ring and conformational structure of the fructose induced by irradiation were identified.58
The mechanism of thermal radical generation in cereal starches with different amylose contents was analysed by using a Raman microspectrometer. Effects of high temperature on the structure of polysaccharide molecules were tracked from the collected Raman spectra. Due to the decomposition of polysaccharide chains by the cleavages of the glycosidic bonds, the highest amount of decrease was observed for the bands at νa (1150 cm−1) and νs (944 cm−1) of C–O–C.59 Different from the previous one, in this study, how freezing treatment affects the structure of wheat bread dough was examined with Raman spectroscopy. The distribution of ice free water, starch, gluten and yeast in the frozen dough in the structure was determined by examining the Raman bands of each of these components, and the microstructure of the dough was determined by making use of images. In this way, researchers stated that the causes of the decrease in quality can be found in the frozen baked goods.60
Lipids
Lipids are one of the three major food components, but they have been reported as the most complex molecular structures to be analysed. Raman spectroscopy has been widely used for determining different properties of lipids. For instance, Sadeghijorabchi et al. put forward a procedure that determines the total level of unsaturation in oils and fats by using FT-Raman spectroscopy.61 Similarly, Silveira et al. quantified unsaturated fats in fat-containing foods. Raman spectra of edible oils, margarine, mayonnaise, hydrogenated fat, and butter were obtained with a near-infrared Raman spectrometer by making use of this non-destructive quantification method. Spectral regions of 1750, 1660, 1440, 1300, and 1260 cm−1 were used to establish a correlation between Raman intensities and the total and unsaturated fat contents of analyzed samples.62 El-Abbasy et al. quantified the fat content in liquid homogenized milk by using VIS-Raman spectroscopy. Protein and carbohydrate content of milk samples didn't made a significant influence on the Raman intensities, so the variations were directly attributed to the fat contents of the samples. Characteristic bands were mostly assigned to the fatty acids and monitored at bands in 1650 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C cis double bond stretching of RHC = CHR), 1440 cm−1 (C–H scissoring of –CH2), 1265 cm−1 (C–H bending at the cis double bond in R–HC
C cis double bond stretching of RHC = CHR), 1440 cm−1 (C–H scissoring of –CH2), 1265 cm−1 (C–H bending at the cis double bond in R–HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–R), 1300 cm−1 (C–H twisting of the CH2 group), and 1747 cm−1 (C
CH–R), 1300 cm−1 (C–H twisting of the CH2 group), and 1747 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching of RC = OOR).63 McGoverin et al. used Raman spectroscopy to quantify milk powder constituents, namely protein and fat in skim and whole milk samples. The overlapped bands seen in Raman spectra are considered to be caused by lactose, milk proteins and milk fats. The characteristic band represented by lower wavenumber 1745 cm−1 C
O stretching of RC = OOR).63 McGoverin et al. used Raman spectroscopy to quantify milk powder constituents, namely protein and fat in skim and whole milk samples. The overlapped bands seen in Raman spectra are considered to be caused by lactose, milk proteins and milk fats. The characteristic band represented by lower wavenumber 1745 cm−1 C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes were assigned to milk fat, while the phenylalanine ring breathing band at 1005 cm−1 was accepted as the indicative of protein. A low broad peak above 3300 cm−1 was reported to be consistent with N–H and O–H modes of protein and lactose.64 A combination of Raman spectroscopy with chemometric methods enabled researchers to establish predictive models for these constituents by using abovementioned characteristic bands. This combination has also been used to predict the abundance of fatty acids in clarified butterfat,65 and to discriminate and classify different oils and fats.66–68 Marquardt et al. used Raman spectroscopy to obtain quantitative data on carotenoid, collagen and fat contents of the fish muscle samples. Fat content was characterized by the bands at 657, 1440, 1301 (CH2 in phase twist), 1267(C–H symmetric rock (cis)), 1076 (C–C–C stretch) and 1064 cm−1 (C–C–C stretch). Carotenoids were monitored at primary bands (1159 and 1518 cm−1), and the intensity of the bands at 857 (proline) and 940 cm−1 (C–C stretch of peptide backbone) which were assigned to the presence of collagen was found to be relatively weak.7
O modes were assigned to milk fat, while the phenylalanine ring breathing band at 1005 cm−1 was accepted as the indicative of protein. A low broad peak above 3300 cm−1 was reported to be consistent with N–H and O–H modes of protein and lactose.64 A combination of Raman spectroscopy with chemometric methods enabled researchers to establish predictive models for these constituents by using abovementioned characteristic bands. This combination has also been used to predict the abundance of fatty acids in clarified butterfat,65 and to discriminate and classify different oils and fats.66–68 Marquardt et al. used Raman spectroscopy to obtain quantitative data on carotenoid, collagen and fat contents of the fish muscle samples. Fat content was characterized by the bands at 657, 1440, 1301 (CH2 in phase twist), 1267(C–H symmetric rock (cis)), 1076 (C–C–C stretch) and 1064 cm−1 (C–C–C stretch). Carotenoids were monitored at primary bands (1159 and 1518 cm−1), and the intensity of the bands at 857 (proline) and 940 cm−1 (C–C stretch of peptide backbone) which were assigned to the presence of collagen was found to be relatively weak.7
Lipid oxidation, one of the most important quality indicators in foods, has been investigated with Raman spectroscopy. Muik et al. examined the chemical changes that occurred during lipid oxidation in edible oils.69 Kathirvel et al. monitored the progression of lipid oxidation in mechanically separated turkey by monitoring the oxidative bleaching of β-carotene using Raman spectroscopy. Three characteristic Raman bands at 1008 cm−1 from the C–CH3 rocking, at 1160 cm−1 from the C–C stretching and at 1524 cm−1 from the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching were observed for β-carotene molecule, while the last one was used to monitor its concentration.70 Guzman et al. determined the oxidation status of olive oil through a combination of low-resolution Raman spectroscopy and PLS analysis. In order to monitor olive oil oxidation, characteristic Raman bands at 1267 cm−1, 1302 cm−1, 1442 cm−1 1655 cm−1, and 1747 cm−1 (corresponding to symmetric rock double bond in cis = CH, in-phase twist methylene, methylene scissoring mode of CH2, cis double bond stretching (C
C stretching were observed for β-carotene molecule, while the last one was used to monitor its concentration.70 Guzman et al. determined the oxidation status of olive oil through a combination of low-resolution Raman spectroscopy and PLS analysis. In order to monitor olive oil oxidation, characteristic Raman bands at 1267 cm−1, 1302 cm−1, 1442 cm−1 1655 cm−1, and 1747 cm−1 (corresponding to symmetric rock double bond in cis = CH, in-phase twist methylene, methylene scissoring mode of CH2, cis double bond stretching (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), and ester stretching (C
C), and ester stretching (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), respectively) were detected in the region below 1800 cm−1.71 In addition to these applications, it is also possible to show the effects of storage conditions on lipids or lipid containing foods. Sanchez-Alonso et al. used FT-Raman spectroscopy to monitor the lipid oxidation of hake fillets during frozen storage. C–C stretching vibration at 1658 cm−1 was reported as the only characteristic Raman band related to the lipid oxidation.72
O), respectively) were detected in the region below 1800 cm−1.71 In addition to these applications, it is also possible to show the effects of storage conditions on lipids or lipid containing foods. Sanchez-Alonso et al. used FT-Raman spectroscopy to monitor the lipid oxidation of hake fillets during frozen storage. C–C stretching vibration at 1658 cm−1 was reported as the only characteristic Raman band related to the lipid oxidation.72
A study was carried out using linoleic acid, which is a very important fatty acid in human diet. In this study, linoleic acid was treated with high pressure. Linoleic acid's phase transition and conformational changes with high pressure were observed in real-time by using Raman spectroscopy. Significant conformational changes were observed at 0.07–0.12 GPa and 0.31–0.53 GPa. With the increase in pressure, some Raman bands disappeared, while some of them appeared. The researchers believe that knowledge about these chemical and physical changes will make the major contribution to food preservation technology.73 Another essential oil produced out of Lamiaceae plant displaying different chemical profiles according to their genomic properties has a biological activity of great importance. In the light of these informations, chemical structures of the essential oils were determined using dispersive Raman spectroscopy and FT-IR. Chemotyping was based on characteristic bands of thymol (740 cm−1 ring vibration) and carvacrol (760 cm−1), and the results were confirmed by using GC.74
Researchers used Raman spectroscopy to follow the changes in carotenoid structure of extra virgin olive oil with heat treatment, which was applied by using microwave and conventional heating processes. It was shown that conventional heat treatment caused more rapid degradation of carotenoid bands at 1008 cm−1 (C–CH3 bend), 1150 cm−1 (C–C stretch), and 1525 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch). In addition, the researchers found that high heat treatment resulted in whole degradation of carotenoids and that application time plays a more important role in the degradation compared to high temperature. They determined that heat treatment with microwave during oil rafination affects the oil quality less than the conventional heat treatment since the desired temperature was achieved by microwave more quickly than conventional heating.75
C stretch). In addition, the researchers found that high heat treatment resulted in whole degradation of carotenoids and that application time plays a more important role in the degradation compared to high temperature. They determined that heat treatment with microwave during oil rafination affects the oil quality less than the conventional heat treatment since the desired temperature was achieved by microwave more quickly than conventional heating.75
Vitamins
A variety of analytical procedures have been used for vitamin analysis in food samples. Lack of specify and matrix effect were reported as the main disadvantages of these procedures. On the other hand, Raman spectroscopy has gained an increasing importance due to its high precision and good signal-to-noise rate for vitamin analysis.76Investigation of vitamins with Raman spectroscopy began in the 1970s. Main goals of these early studies were to characterize the isomeric forms77 and obtain the characteristic Raman spectra of vitamins.78 Rimai et al. acquired the Raman spectra of retinals (trans, 9-cis, 13-cis), retinols (trans, 13-cis), and trans-retinoic acid in octanol solution and reported the possibility of characterizing the terminal group on vitamin A type molecules and isomers by their characteristic bands at around 1580–1590 cm−l and 1100–1400 cm−l.77 Similarly, Tsai and Morris researched the effect of pH and other water soluble vitamins on Raman intensity of Vitamin B12 by using cyanocobalamin as a model chemical. A strong Raman band at 1504 cm−1 corresponding to the ring stretching vibration of molecule was followed.79 Also, Cimpoiu et al. coupled high performance thin layer chromatography (HPTLC) with Raman spectrometry in order to obtain a suitable method for identification of eight hydrophilic vitamins, i.e., B1-thiamin, B2-riboflavin, B3-nicotinic acid, B5-panothenic acid, B6-pyridoxine, B9-folic acid, B12-cyanocobalamin, and C-ascorbic acid in different samples. In this study, a successful separation was achieved by HPTLC, and vitamins were easily characterized by Raman spectroscopy.80
The use of Raman microscopy also made it possible to determine and localize vitamins in biological samples. Kim and Carey used riboflavin to differentiate free vitamins and vitamins bound to vitamin binding proteins at micro molar concentrations.81 In another study, Beattie et al., used Raman spectroscopy to identify α-tocopherol, which is known to be the predominant form of vitamin E in biological samples.82
Additionally, chemometric techniques were used in order to quantify vitamins in powdered mixtures and solutions. Spectral regions between 2800–3000 cm−1 and 800–1750 cm−1 were used for the PLS models due to their high correlation with Vitamin C concentrations. A detailed chemical assignment was given in the relevant study.83
Raman spectroscopy for microorganism and virus detection
There are several analytical methods to determine the presence of microorganisms and viruses. Although traditional microbiological plate count methods such as PCR, and immunological and serological methods have been frequently used for this purpose, Raman spectroscopy has gained increasing attention due to its abovementioned advantages such as high sensitivity, reliability and non-destructiveness.84 In addition to microorganisms that grow in food, Hepatitis A and Norwalk viruses, Poliovirus, Astrovirus, Enteric adenovirus, parvovirus and rotaviruses have also been found in foods as contaminants.84,85 Specific Raman bands for microorganism and virus detection were given in Table 5S.†Microorganisms
Raman and its derivatives, such as UV-RR, FT-Raman, micro-Raman and confocal Raman can be used to determine the presence of microorganisms. By carefully selecting which Raman method to use, taking advantage of neural networks, and utilizing chemometric methods to make qualitative distinctions between spectra; it is possible to identify and differentiate microorganisms.86,87Bacillus and Brevibacillus species are spore-forming bacteria. Bacillus species, in particular, is pathogenic and causes serious food poisoning incidences; they can also be used as a biological weapon. Passage from the spore to the vegetative forms of these bacteria, and determining the effects of manganese dipicolinate and calcium dipicolinate upon spore formation, were monitored with micro-Raman spectroscopy.88 In a study made on spore-forming Clostridium cultures, single-cell spectra were collected using confocal Raman microscopy. Although the morphological structures of the cells were similar, significant spectral differences were observed between them and their contents depending on age and spore production. As a result, chemical differences between the cells were easily identified by Raman microscopy.52
In another study with confocal Raman microscopy, a group of pathogenic microorganisms (Enterococci and Staphylococci) was classified. Raman measurements were taken from different regions of a microcolony of each culture and processed with chemometric methods. When dendrograms obtained with Hierarchical Cluster Analysis (HCA) were analysed, two arms were observed, one of which belongs to Enterococci and the other to Staphylococci.89 Specific strains of Staphylococcus were identified with a micro-Raman system. With this methodology, it was possible to determine chemotaxonomic classification for a single cell and bulk cultures. HCA and support vector machine (SVM) were used as statistical methods. In one of the analyses, all of the bacteria were incubated under the same conditions (medium type, incubation temperature and time), and their Raman spectra were taken. Then, the effect of changes in culture media, incubation temperatures and times were also analyzed.90 Another research carried out with Bacillus and Brevibacillus species covered identification and differentiation of these bacteria with a UV-RR technique. The researchers reaffirmed the accuracy of their results with analyses of 16S rDNA of the bacteria. They stated that genotypic and phenotypic differences between microorganisms could be detected by using characteristic Raman bands obtained from cellular components such as aromatic amino acids and UV adsorption-capable nucleic acids. The spectra were obtained at a wavelength of 244 nm, and subjected to multivariate statistical methods.91 By using FT-Raman system, Yang and Irudayaraj separated six different microorganisms (S. cerevisiae, Fusarium verticillioides, Bacillus cereus, Aspergillus niger, Escherichia coli, L. casei) from each other as well as different strains belonging to the same species. They reported differences due to cell structures within a ‘fingerprint’ range of 600–1800 cm−1 for the microorganisms. PCA and CVA were used to characterize these microorganisms.87 In another study by Maquelin et al., FT-IR and FT-Raman spectra were conducted on dehydrated Enterococcus faecalis. C–H stretching bands belonging to (CH3, CH2, and CH) functional groups were observed in the 2700–3000 cm−1 region, and the deformation band of the C–H bond at 1450 cm−1 was also present. Protein amide I and amide II bonds and vibrations of bases in RNA/DNA were also detected.92 Colonies of Microccoccus luteus (M. luteus), Bacillus subtilis (B. subtilis), Pseudomonas fluorescens (P. fluorescens), Rhodotorula mucilaginosa (R. mucilaginosa), and Bacillus sphaericus bacteria were analysed with FT-Raman. It was found that the spectra of M. luteus, B. subtilis and P. fluorescens had completely different spectra from each other at a wavelength of 785 nm incident light. As a result of stimulation with light at 633 nm, distinct templates were obtained from cells belonging to the pigmented bacteria M. luteus and R. mucilaginosa.86 The previously mentioned group also separated twenty different Micrococcus, Bacillus, E. coli and Staphylococcus strains using micro-Raman spectroscopy coupled with SVM as chemometric analysis.90 Raman spectroscopy coupled with different chemometric methods was also used for the identification of Legionella, Klebsiella, Micrococcus, Bacillus, E. coli, Pseudomonas, Staphylococcus, Listeria, Yersinia and Salmonella species.93–97
Micro-Raman spectroscopy was also used to distinguish different types of Lactarius mold by using chemometric methods. Lipid and amylopectin were monitored with Raman spectroscopy since these are characteristic compounds for amyloidal reactions of Lactarius spores. Lactarius mold is of great importance in ecological and economic sense and popular in many regions of the world owing to its being edible.98 Micro-Raman spectroscopy has also been used to investigate the spatial distribution and composition of lipid vesicles inside intact hyphae of Mortierella species. Differences in the degree of unsaturation and the effect of growth conditions on lipid composition were determined for Mortierella alpina and Mortierella elongata species.99
Viruses
There have been many studies reporting the use of Raman spectroscopy for structural characterization of viruses. For instance, a number of studies have been conducted using structural information obtained out of Raman spectroscopy to develop antiviral drugs.100–103 In one of these studies, the formation mechanism of the icosahedral capsid of P22 phage, which is effective against Salmonella typhimurium, was tracked using a Raman microdialysis flow cell. After preparation of the procapsid, empty shell, and scaffolding protein of the phage, all the components were placed in the Raman microdialysis flow cell system and their Raman signals were collected. The results were verified with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and the CD spectroscopy. In accordance with their conclusions, researchers have managed to develop models for the transformation of procapsid into capsid, and procapsid assembly.102 UV-Raman has been used to investigate the protein structure of another phage. Raman spectra of the phage were obtained by excitation at four different wavelengths (257, 244, 238 and 229 nm). As a result of excitation at 257 nm, signals of the bases that build the genome were obtained, and characteristic Raman bands corresponding to the amino acids of the coat protein were attained as a result of excitation at 229 nm.103,104 Another research group has benefited from Raman optical activity (ROA) for the structural characterization of nucleic acids, viruses and proteins in a manner distinct from other studies. The researchers found that working even with the full virus was possible, and information on both the coat proteins, and the nucleic acids enclosed in the capsid could be obtained.100,101There are very few studies on the analysis of foodborne viruses by using Raman spectroscopy. Actually, there is a single study using Raman spectroscopy on Hepatitis A, which is the most common foodborne virus.105 Hepatitis A 3C proteinase is known to be a cysteine protease which is very important for the life cycle of this virus and responsible for the formation of mature viral proteins from the polyprotein precursor.106 In the aforementioned research, Raman spectra were used to investigate acyl groups in the active site of the enzyme.105 There have been several studies utilizing Raman spectroscopy for the investigation of Hepatitis viruses, but none of them have been reported as a foodborne virus.107–109
Raman spectroscopy for toxin and chemical detection
Contaminants are substances that have not been intentionally added to food. These substances may be present in foods as a result of contamination in any stage of the production, packaging, transport or storage. They can also result from the environmental contamination. Since contaminants in general have a negative impact on the quality of food and are a threat to human health,110 a number of analytical methods have been developed for the identification and quantification of these compounds. Raman spectroscopy is one of these methods which has gained increasing attention in recent years. Specific Raman bands related to toxin and chemical detection were given in Table 6S.†Toxins
Brandt et al. aimed to study the structural properties of the toxins ricin, ricin agglutinin and ricin binding subunit B. ricin and ricin agglutinin were extracted from Ricinus communis seeds and purified using affinity chromatography and gel-filtration. Vibrational bands of this plant toxin were then obtained by using Raman spectroscopy. Amide I at 1640 cm−1, amide III at 1210–1300 cm−1, a tyrosine doublet at 830 and 855 cm−1, bands for disulphide bridges at 510, 525 and 540 cm−1, and some several bands corresponding to tryptophan amino acid residues at 1361 cm−1 were used as conformation sensitive bands for the molecules of interest.111In another study, several vegetables and fruits were analysed with micro-Raman and near-infrared FT-Raman spectrometry to detect trace amounts of residual pesticides on the surface.112 Bonora et al., investigated the Raman spectra of atrazine, prometryn and simetryn herbicides in solid form and in polar and apolar solvents. A comparison was made between theoretical spectra and experimental spectra obtained from Raman and surface-enhanced Raman spectroscopy (SERS) measurements.113 In a similar study, Fleming et al. investigated the molecular structure of phosphorus-containing herbicides. IR, Raman and SERS spectra were collected and compared with density functional theory (DFT) calculations.114
Deoxynivalenol (DON) is one of the major secondary metabolites of the Fusarium genus and found predominantly in grains such as wheat, barley and corn.115 The presence of DON degrades the quality of grain and has toxic effects on human health.116 Traditional methods to measure DON concentrations in grain involve time-consuming steps such as extraction, washing and binding.117 Due to the high moisture content in grain, broad intense water bands are yielded in both the IR and NIR regions. Thus, highly informative bands attributable to carbohydrate and protein species are inhibited. For this reason, various kinds of studies have been conducted with IR spectroscopy. The only study in the literature for identification of DON toxin by using a Raman technique with infrared spectroscopy was published by Liu et al. In this study, feasibility of FT-Raman spectroscopy for the characterization and classification of ground wheat and barley contaminated with varying amounts of DON was investigated. PCA was performed in the spectral region 1800–800 cm−1 for multiplicative scatter correction of the Raman spectra. Principal component scores were then examined to discriminate between low and high DON in wheat.118
Raman spectroscopy coupled with LDA was used for qualitative and quantitative analysis of aflatoxin produced by Aspergillus in maize. Differences in the Raman bands were observed depending on the aflatoxin concentration in the samples.119 In another study carried out by Lee et al., three different vibrational spectrophotometric methods, namely Raman, FT-NIR, and FT-IR were used for the detection of aflatoxin in different concentrations. By applying different chemometric methods to the spectra obtained from these three methods, a classification was made according to their aflatoxin quantities. The researchers stated that based on the results of the chemometric method they applied, Raman and FT-IR analyses had given relatively more satisfactory results compared to FT-NIR.120
In a study by Gupta et al., in situ synthesis of a nanopatterned conjugated molecularly imprinted polymer for bioagent T-2 on a bare gold chip and its integration with surface plasmon resonance and Raman spectroscopy were explored. The p-aminophenylboronicacid (p-APBA) and p-APBA with T-2 were characterized with Raman spectroscopy. Upon polymerization of 3-APBA with T-2, the presence of new bands was detected, and they were assigned to symmetric B–O and asymmetric C–O stretching modes for p-APBA and T-2 in the sample.121
Chemicals
Coumarin is a naturally occurring benzopyrone found in most plants including tonka beans, sweet clover, woodruff and grass. It was used as a flavouring food additive until its direct use was banned due to the concerns about hepatotoxic effects on animal models.122 Sortur et al. reported that IR and Raman spectra of 6-methyl-4-bromomethylcoumarin were obtained by following the reaction of p-cresol with 4-bromoethyl acetoacetate on an ice bath.123Bisphenol A (BPA) is an estrogenic compound widely used in polycarbonate plastics, food cans and food storage containers.124 Dybal and co-workers prepared BPA samples by varying thermal and solvent treatments. Characteristic amorphous bands at 735 and 1235 cm−1 were used to determine the degrees of crystallinity of the BPA polycarbonate samples.125
Ground waters may be contaminated with perchlorate ions originating from the use of fertilizers and manufacturing activities. Levitskaia, Sinkov and Bryan used perchlorate loaded ion exchange resin as their model system. Determination of perchlorate with Raman spectroscopy was found to be a practical real time detection method. Perchlorate, a tetrahedral anion possessing easily polarizable Cl–O bonds, exhibits a vibrational frequency of the symmetric stretch near 934 cm−1 in aqueous solution. The ClO4− bands were normalized to the intensity of the prominent A850 resin band at 1452 cm−1 that served as an internal standard.126 Yu et al. used RR spectroscopy for quantitative analysis of divalent metal ions.127 Chelation property of zincon molecule with Cu2+ and Ni2+ enabled researchers to obtain complexes which could be followed by RR spectroscopy.
Polycyclic aromatic hydrocarbons (PAH) constitute a potential health danger because of their ability to induce carcinogenesis. PAH (such as naphthalene, anthracene, phenanthrene, and pyrene) could be detected in trace levels by making use of UV-RR spectrometry. A strong band for naphthalene was located at 766 cm−1. Other strong bands were observed at 399, 756 and 1407 cm−1 for anthracene, at 386, 745 and 1386 cm−1 for phenanthrene and at 582, 1393, and 1622 cm−1 for pyrene.128 Alajtal, Edwards and Scowen used FT-Raman spectroscopy to investigate the effect of spectral resolution on the Raman spectra of several polyaromatic hydrocarbons. In this study, Raman measurements of beta-carotene naphthalene, β-carotene anthracene, β-carotene pyrene, and naphthalene, anthracene, and pyrene molecules were taken with different spectral resolutions. The effect of spectral resolution on the obtained Raman spectra was evaluated in this study.129
In the study of Sundaraganesan, Puviarasan and Mohan, the vibrational spectra of acrylamide found in starchy food products as a result of cooking practices was discussed in detail with respect to various environments. A complete vibrational assignment using polarization data along with the results of normal coordinate analysis were presented in this study by taking into account the internal modes of the CH2 and NH2 groups.130
Raman spectroscopy in food additive analysis
Raman spectroscopy has also been used to detect food additives, and different approaches have been taken into account for this purpose. Specific Raman bands for food additive analysis section were given in Table 7S.†Astaxanthin (E-161j) and canthaxanthin (E-161g), the two major carotenoids responsible for the red-orange colour of salmon, were investigated by using Raman spectroscopy. Strong Raman signals were observed as a result of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretch vibrations of the carotenoid molecules.131 Carbon black (E-153), another colouring agent which is produced by the combustion of hydrocarbons, was analysed with Raman spectroscopy.132 Snehalatha et al., investigated the molecular structure of amaranth (E-123), a commonly used colouring agent of the food industry. Most characteristic bands were assigned to the vibrations of naphthalene ring and azo chromophoric group (C–N
C stretch vibrations of the carotenoid molecules.131 Carbon black (E-153), another colouring agent which is produced by the combustion of hydrocarbons, was analysed with Raman spectroscopy.132 Snehalatha et al., investigated the molecular structure of amaranth (E-123), a commonly used colouring agent of the food industry. Most characteristic bands were assigned to the vibrations of naphthalene ring and azo chromophoric group (C–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–C). A medium intensity band in the Raman spectrum was identified as the symmetric stretching vibration of the SO3 group. The strongest Raman band was obtained out of the naphthalene ring vibrations.133 Peica et al. studied the molecular structure of tartrazine (E-102), an artificial dye which is also known for its potential to cause allergic reactions. Its strongest bands resulted from the azo and carboxyl groups and C–H deformation of the phenyl groups.134 Curcumin is a natural coloring agent and stabilizator in the food industry as it is the major contributor to human health, yet it has a limited application area because of its low solubility and stability. To enhance solubility and stability of curcumin, encapsulation was applied with cyclodextrin, and then characterization of this complex was accomplished with Raman spectroscopy. The researchers showed that Raman spectra of curcumin–cyclodextrin complex were different from Raman spectrum of curcumin.135
N–C). A medium intensity band in the Raman spectrum was identified as the symmetric stretching vibration of the SO3 group. The strongest Raman band was obtained out of the naphthalene ring vibrations.133 Peica et al. studied the molecular structure of tartrazine (E-102), an artificial dye which is also known for its potential to cause allergic reactions. Its strongest bands resulted from the azo and carboxyl groups and C–H deformation of the phenyl groups.134 Curcumin is a natural coloring agent and stabilizator in the food industry as it is the major contributor to human health, yet it has a limited application area because of its low solubility and stability. To enhance solubility and stability of curcumin, encapsulation was applied with cyclodextrin, and then characterization of this complex was accomplished with Raman spectroscopy. The researchers showed that Raman spectra of curcumin–cyclodextrin complex were different from Raman spectrum of curcumin.135
Zborowski et al., used IR and Raman spectroscopy for characterizing the molecular structure of maltol which is widely used as a natural food additive. Maltol was characterized with its strong band related to the O–H stretching.136 Peica et al. used Raman spectroscopy to investigate the molecular structure of monosodium glutamate which is a commonly used flavour enhancer in various food products. Strong Raman bands were explained by CH2 stretching, COO− stretching, CH2 deformation, completely ionized form, and COO− twisting.137 In another study by Peica et al., aspartame (E-951) as an artificial sweetener was analysed by using Raman spectroscopy. Strong Raman bands were assigned to symmetrical C–H phenyl ring stretching, in-plane C–H phenyl ring bending, symmetrical stretching, phenylalanine ring stretching, CH3 rocking, and skeletal deformation.138
Potential of IR, Raman and SERS to determine the excess azodicarbonamide additive in flour samples was evaluated. Its reaction products, namely biurea and semicarbazide that were formed during baking process were monitored by following their characteristic Raman spectra. Although multiple characteristic Raman bands were observed for each product, their presence is mostly assigned to the deformation bands of NH2, stretching and bending vibrations of N–C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bounds. Results taken from experimental and calculated Raman spectra were verified with DFT.
O bounds. Results taken from experimental and calculated Raman spectra were verified with DFT.
Another food additive, chitosan, which is obtained by deacetylation of chitin, is of great importance since the degree of its deacetylation is vital in order to determine its chemical and physical properties such as solubility, biodegradability and biocompatibility.139 In this respect, Zajac et al. demonstrated that the deacetylation degree of chitosan could be calculated by following certain bands obtained from Raman and IR spectra related to it.140 The other chitosan derivative obtained by sulfating are known to have anticoagulant, antiviral, antimicrobial, and antioxidant characteristics. It could be possible to determine the degree of substitution and to characterize sulfated chitosan compounds using Raman spectroscopy. Changes due to binding of sulfate groups to chitosan were detected in the obtained Raman spectra (1070, 1014, 823–834, 580–610, 2964 cm−1) and these compounds were characterized according to the amount of the sulfate group attached to the structure.141
Another food additive is mannitol, which is used in the production of low-calorie food, as well as in the pharmaceutical industry and in other lyophilized products. In a study, changes occurring in the bands of ice, water and mannitol during the lyophilization of mannitol, were examined. Raman bands of ice and mannitol were monitored at the spectral regions 150–250 cm−1 and 1000–1170 cm−1, respectively. Different polymorphic forms of mannitol were displayed during the lyophilization process.142
Raman spectroscopy in raw material analysis
Rapid and in situ analysis of raw materials is one of the most important quality control applications in food industry. Assessing the quality of raw materials before the food production phase helps manufacturers save time and reduce the cost. Identification of raw materials is also essential due to its major effect on the quality of final product. Taking these requirements into account, it can be stated that Raman spectroscopy provides a wide range of application area in which the raw material analysis constitutes a significant part. In this context, Raman spectroscopy has been widely used for raw material analysis, particularly for the discrimination of food samples, monitoring chemical and biochemical processes, compositional characterization of food samples and authentication of foods. Nevertheless, analyzing the Raman spectra of food samples mostly requires chemometric tools because of the complex structure of food matrix. Unsupervised chemometric methods like principal component analysis (PCA) and supervised chemometric methods like partial least square (PLS), partial least square-discriminant analysis (PLS-DA), principal component regression (PCR) as well as artificial neural networks (ANN) were generally employed for the detailed analysis of collected Raman data.143–146 Specific Raman bands reported in the context of the reviewed articles is given in Table 8S† for raw material analysis.Honey
In a study by Goodacre, Radovic and Anklam, Raman spectroscopy was used with PCA and ANN to discriminate between honey samples provided from various European countries with different floral and geographical origins.144 First, scores representing the Raman spectra of honey samples were obtained and then ANN, which was created from these scores, was used for discrimination. According to the results, 13 of 14 honey samples were classified accurately, but the country of origin was not predicted successfully as the number of honey samples was insufficient. Another research on honey has also recently been carried out by Özbalci et al. In this study, sugar contents of honey samples were quantified by applying chemometric methods to Raman spectra of honey samples.147 Similar to the first study mentioned in this review, Carvucci et al. discriminated the honey samples collected from different regions by using Raman spectroscopy. They processed the Raman spectra that they obtained according to the pollen composition of genuine honey by using PCA, and identified the botanical and geographical origins of it.148Coffee
There are three studies in the literature conducted for discriminating between Arabica and Robusta green coffee by using Raman spectroscopy.143,149,150 The reason for the researchers to discriminate these two kinds coffee is that their quality, and thus price is not the same. Analyzing their lipid content was the focus of these studies, and it was found that especially their kahweol content considerably differs from each other. The first Raman study on this topic of interest was carried out by using FT-Raman spectroscopy. The researchers obtained Raman spectra of lipid samples (kahweol and cafesto) extracted from coffee samples. Two characteristic peaks (1567 vs. 1478 cm−1) of kahweol were found in the extract taken from Arabica coffee, which is specific to this type of coffee. They also discriminated between these two coffee types with a success rate of 93% by using chemometric method PCA.150 Similarly, in a subsequent study, these two kinds of coffee were discriminated by using FT-Raman. However, it differs from the former study in that Raman spectra of the samples were taken without applying any chemical and physical procedure on the coffee beans. The discrimination of the coffees was performed by calculating the “spectral kahweol index” with the spectra obtained from the samples with different geographical origins.149 In another study, chlorogenic acid rate was also examined as well as the lipids present in Arabica and Robusta green coffee. Raman spectra of the samples were obtained with visible micro-Raman spectroscopy; and by using two different PCA models, the discrimination of the coffees were accomplished with a success rate of 93%.143Lipid
A study on discrimination among different edible oils and fats was performed by Yang et al. In this study, spectra obtained from FT-Raman spectroscopy were compressed with PLS and PCA; then the processed data were used for linear discriminant analysis (LDA) and canonical variate analysis (CVA). As a result of the analyses performed using the spectral range between 400 and 3700 cm−1 (Table 1S†), PLS-CVA was found to be the best method for discriminating edible oil and fats by FT-Raman, with calibration and validation data of 93.3% and 94.4%, respectively.68 Korifi et al. tried to evaluate the capability of confocal Raman spectroscopy combined with chemometric treatments to authenticate virgin olive oils with the protected designation of origin (PDO) label. Hence, PLS-DA was applied to the spectra of eight French PDOs, and 92.3% of these oils were accurately classified.151 Raman spectroscopy has also been used to determine the quality of the olive fruit, which is one of the most important quality parameters in olive oil processing. In another study on olive oil, it was put forward that low-resolution portable Raman system could be utilized for determining oxidation states of oils. Having benefited from a chemometric method (PLS), the researchers stated that due to oxidation, changes occurred in the bands at about 1267, 1302, 1442, 1655 vs. 1747 cm−1.71 They also tried to determine the quality of olive oil by using the Raman spectra of ground or sound olive paste, and then attempted to discriminate the origin of the olives as ground or sound. In the first stage of the study, PCA was used to find natural clusters in the Raman spectra, and then supervised classification methods, namely Soft Independent Modelling of Class Analogy (SIMCA), PLS-DA and K-nearest neighbors' (KNN) were applied. The best results for classification were found in the KNN method, with prediction abilities of 100% for sound and 97% for ground olives in an independent validation set. In this study, it was demonstrated that portable Raman spectroscopy can be utilized to determine the quality of olives used in the production of olive oils in the field.152 Different from the other studies on olive oil, Gouvinhas et al., produced extra virgin olive oil by means of taking samples from three types of olive in different stages of their ripening periods. They were classified according to their types and ripening periods through processing the Raman spectra with qualitative methods. It was found that 1749, 1651, 1439, 1303 vs. 1267 cm−1 bands obtained from Raman measurements directly demonstrates the fatty acid contents of the samples, and changes were observed in the intensities due to ripening.153In another study, different types of pure animal fat (poultry, pig, bovine, and lamb and fish oils) and mixture samples of them were classified according to their origin by using PCA and PLS-DA analyses.66 The same analysis was performed with different types of edible fats. Fish, poultry, pig and bovine fats were well distinguished by applying PCA. Using PLS-DA analysis, poultry, pig and bovine fat samples were discriminated with high sensitivity and specificity values and with few classification errors. In addition, types of edible fats like fish oils and acidic oils from chemical or physical refining could also be discriminated with PLS-DA.66 Velioğlu et al., used Raman spectroscopy to assess the freshness of fish samples according to the number of freezing–thawing cycles they were exposed to. PCA was employed to cluster the samples according to their freshness. Changes in the intensities of the characteristic Raman bands were mostly attributed to the alterations in the lipid structure.154 Potential of Raman spectroscopy to predict the purity of caviars was evaluated. Linear methods such as PCA and LDA as well as non-linear methods such as ANN were used to classify different caviar samples according to their purity and type. More accurate predictions were obtained by using the ANN with 91.4% of prediction capability. Fatty acids and fat contents of the caviar samples was quantified through Raman spectroscopy coupled with PLS regression.155
In addition to the abovementioned studies on lipids, there is another study in which Raman spectra of 35 lipids belonging to different families (saturated and unsaturated fatty acids, triacylglycerols, cholesterol, cholesteryl esters and phospholipids) were obtained. It was found that Raman spectra of each of these lipids display changes depending on their saturation state, their being in liquid and solid state, and on isomer forms. The characteristic features of Raman spectra is attributable to the existence of hydrocarbon chains, and they were observed at 1500–1400, 1300–1250, 1200–1050, 3000–2800 cm−1, respectively, which was caused by C–C and C–H stretching modes and the scissoring and twisting vibrations of CH2 and CH3 groups. Besides, lipids belonging to each group was found to have characteristic bands specific to them.156
Fermentation products
Raman spectroscopy can also be used to detect materials such as ethanol, lactic acid, and acetic acid that are produced as a result of processes like fermentation. Sivakesava et al., followed ethanol fermentation by Saccharomyces cerevisiae (S. cerevisiae) using Fourier transform-middle infrared (FT-MIR) and FT-Raman spectroscopy. In this study, quantities of glucose, ethanol and the optical cell density of S. cerevisiae during fermentation were investigated using chemometric methods.157 In another study, FT-MIR, Fourier transform-near infrared (FT-NIR) and FT-Raman spectroscopy were used during lactic acid fermentation to determine quantities of the same parameters as the previous study of Lactobacillus casei (L. casei).158 Similarly, quantitative measurements of glucose during ethanol fermentation in the beverage industry were carried out by Delfino et al.37 Our research group has also monitored a two-step acetic acid fermentation in a study using Raman spectroscopy. The first step was consumption of sugars in a grape juice mixture and then formation of alcohol by S. cerevisia. The second step was carried out with Acetobacter aceti that converted the alcohol to acetic acid.159 Wang et al. used Raman Spectroscopy to monitor the consumption and formation of glucose, glycerol and ethanol during wine fermentation. HPLC was used for the validation analysis.160 Micro-Raman spectroscopy was used to follow the fermentation process during yoghurt production. Chemical transformation of lactose and inorganic phosphorus into lactic acid and organic phosphorus and the formation of the exopolysaccharides were monitored based on the collected Raman spectra as a function of the incubation time.161Other foods
A dispersive Raman spectroscopic method was developed to determine protein and oil contents of soybeans.162 Optimal prediction models were generated by PLS algorithms based on collected Raman spectra (200–1800 cm−1) of the samples. Protein and oil content of the soybeans were successfully predicted with high R2 values (0.916 and 0.872 for protein and oil contents, respectively). Characterization of foods is another topic studied by using Raman spectroscopy. For this purpose, FT-Raman spectroscopy was used for the characterization of Marama beans from Southern Africa. Both quantitative and qualitative data on the composition of Marama bean oil, including carbohydrates, proteins, amino acids and aromatic compounds, were obtained.145 Ripe and unripe tomato fruit samples were analyzed with portable and confocal Raman microscope to obtain spectral data on their main organic components. Two different laser excitation wavelengths were used for confocal microscope measurements to maximize the obtained spectral information.163 By using spectral data, cutin and cutinal waxes on unripe tomatoes and carotenes, and polyphenols and polysaccharides on ripe tomatoes were identified as major compounds. In another study, the researchers traced the lycopene formation and distribution in the structures of the harvested tomatoes during different stages (green, breaker, turning, pink, light red, red) of their ripening period by using Raman chemical imaging. Tomatoes in different ripening periods were cut parallel to the plane, and Raman spectra were taken from their seeds, locular tissues and outer pericarps. As a result of the trials, two basic peaks (1151 and 1513 cm−1) belonging to lycopene were detected both in locular tissues and on outer pericarps of fully ripened (red) tomatoes.164 Gonzalves et al., used transmission resonance Raman spectroscopy to investigate the spatial distribution of carotenoids in carrot roots, and they found that the changes in the intensities of Raman bands obtained from different parts of carrots were attributed to molecular configuration of β-carotene. As a consequence, β-carotene showed a heterogeneous distribution, and seen particularly in the secondary phloem tissue, and periderm.165Raman spectroscopy can also be used to monitor important components such as ethanol, lactic acid, and acetic acid produced during fermentation166,167 and/or spoilage of foods168 and chemical and biochemical transformations.169,170
Raman spectroscopy to detect food adulteration
Food is adulterated by unscrupulous producers in order to benefit economically from falsifying food information. The development of new techniques to verify food safety and authenticity has been an important issue thanks to the increasing consumer awareness.1 For this reason, rapid and eco-friendly techniques have replaced time-consuming and tiresome chemical and traditional reference methods. As a vibrational technique, Raman spectroscopy is one of the analytical tools and is attracting growing attention due to its ability to provide fingerprint characteristics of food products and its offering a rapid, non-destructive and cheap analysis. In addition, quantitative and qualitative information can be obtained from a combination of Raman spectroscopy with multivariate data analyses.171 Specific Raman bands of related articles to this section were given in Table 9S† for food adulteration.Zou et al. used a portable Raman spectroscopy to distinguish between genuine olive oil and the oil adulterated with low quality oils. A method was developed based on the normalization of cis-(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H) and cis-(C
C–H) and cis-(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) bands intensities at 1265 cm−1 and 1657 cm−1, by the CH2 band intensity at 1441 cm−1. Adulterated olive oil containing as little as 5% (v/v) or more of other edible oils have been successfully detected in the relevant study.172 Zhang et al. investigated extra virgin olive oils adulterated with soybean, corn or sunflower seed oil by characterizing their Raman spectra in the 1000–1800 cm−1 range. The Raman spectra were normalized according to the CH2 band of the oil samples. An external standard method (ESM) was applied to achieve quantitative analysis and compared with the results of SVM methods. Potential of ESM based on Raman spectroscopy to detect olive oil adulteration was shown in this study.173 In another study of Zhang et al., the level of adulteration in a set of olive oil samples containing 5% or more of different types of oils such as soybean, rapeseed, sunflower and corn oil was successfully determined. Using PCA made it possible to obtain a clear separation of oil samples according to their different mono-unsaturated fatty acid, polyunsaturated fatty acid, and saturated fatty acid contents.174 Lopez-Diez et al. also investigated the authentication of various extra virgin olive oils, and their adulteration with hazelnut oil by using Raman spectroscopy. The obtained Raman spectra were normalized according to the frequency of the band representing the scissoring-bending mode of –CH2 groups. The spectra were examined by using PCA, PLS and genetic programming. Extra virgin olive oils from different parts of the Italian peninsula and their mixtures with hazelnut oils were characterized using PCA. The PLS method was also used as a predictive linear model.175 El-Abassy et al. studied visible Raman spectroscopy to classify different vegetable oils and quantify the adulteration of virgin olive oil with sunflower oil. PCA was used for the classification study, while PLS regression analysis was used to monitor the adulteration. Quantitative detection limit was decreased to 500 ppm (0.05%), which is significant in the case of allergic reactions.176 Adulteration of extra virgin olive oil with olive pomace oil was determined by means of NIR, FT-IR and FT-Raman spectroscopy. PLS was used to quantitatively analyse the olive oil samples adulterated with different rates of olive pomace oil.177 Baeten and Aparicio conducted European project FAIR-CT96-5053, which evaluated the performance of FT-Raman, NIR and FT-MIR spectroscopy to authenticate one-hundred thirty-eight different edible oil and fat samples.178 PCA and stepwise linear discriminant analysis (SLDA) methods were performed to classify oils and fats by conducting cluster and discriminant analyses. Three clusters of samples that were rich in saturated fatty acids, monounsaturated fatty acids or polyunsaturated fatty acids were obtained with PCA according to the degree of unsaturation of the oils. In another method, SLDA was performed to classify edible oils and fats according to their sources, and to quantify virgin olive oil adulteration. In one of the studies reported by our research group, Raman spectroscopy was used to detect the adulteration in butter samples spiked with margarine. Prediction success of the models created by using different chemometric methods namely PCA, PCR, PLS and ANN were compared.179
C) bands intensities at 1265 cm−1 and 1657 cm−1, by the CH2 band intensity at 1441 cm−1. Adulterated olive oil containing as little as 5% (v/v) or more of other edible oils have been successfully detected in the relevant study.172 Zhang et al. investigated extra virgin olive oils adulterated with soybean, corn or sunflower seed oil by characterizing their Raman spectra in the 1000–1800 cm−1 range. The Raman spectra were normalized according to the CH2 band of the oil samples. An external standard method (ESM) was applied to achieve quantitative analysis and compared with the results of SVM methods. Potential of ESM based on Raman spectroscopy to detect olive oil adulteration was shown in this study.173 In another study of Zhang et al., the level of adulteration in a set of olive oil samples containing 5% or more of different types of oils such as soybean, rapeseed, sunflower and corn oil was successfully determined. Using PCA made it possible to obtain a clear separation of oil samples according to their different mono-unsaturated fatty acid, polyunsaturated fatty acid, and saturated fatty acid contents.174 Lopez-Diez et al. also investigated the authentication of various extra virgin olive oils, and their adulteration with hazelnut oil by using Raman spectroscopy. The obtained Raman spectra were normalized according to the frequency of the band representing the scissoring-bending mode of –CH2 groups. The spectra were examined by using PCA, PLS and genetic programming. Extra virgin olive oils from different parts of the Italian peninsula and their mixtures with hazelnut oils were characterized using PCA. The PLS method was also used as a predictive linear model.175 El-Abassy et al. studied visible Raman spectroscopy to classify different vegetable oils and quantify the adulteration of virgin olive oil with sunflower oil. PCA was used for the classification study, while PLS regression analysis was used to monitor the adulteration. Quantitative detection limit was decreased to 500 ppm (0.05%), which is significant in the case of allergic reactions.176 Adulteration of extra virgin olive oil with olive pomace oil was determined by means of NIR, FT-IR and FT-Raman spectroscopy. PLS was used to quantitatively analyse the olive oil samples adulterated with different rates of olive pomace oil.177 Baeten and Aparicio conducted European project FAIR-CT96-5053, which evaluated the performance of FT-Raman, NIR and FT-MIR spectroscopy to authenticate one-hundred thirty-eight different edible oil and fat samples.178 PCA and stepwise linear discriminant analysis (SLDA) methods were performed to classify oils and fats by conducting cluster and discriminant analyses. Three clusters of samples that were rich in saturated fatty acids, monounsaturated fatty acids or polyunsaturated fatty acids were obtained with PCA according to the degree of unsaturation of the oils. In another method, SLDA was performed to classify edible oils and fats according to their sources, and to quantify virgin olive oil adulteration. In one of the studies reported by our research group, Raman spectroscopy was used to detect the adulteration in butter samples spiked with margarine. Prediction success of the models created by using different chemometric methods namely PCA, PCR, PLS and ANN were compared.179
Adulteration of milk powder with melamine was determined quantitatively by using portable Raman spectroscopy coupled with PLS regression. Melamine adulteration was monitored using the characteristic bands of melamine located at one strong band at 673 cm−1 and a weak band at 982 cm−1. The intensity of the band at 673 cm−1 was used for quantification of melamine concentration in milk powder.180 In a similar study, adulteration of milk powder with melamine was examined using the melamine band located at 676 cm−1.181 Adulteration was successfully determined in the milk powder samples spiked with calcium carbonate. Prominent peak of calcium carbonate located at 1085 cm−1 was followed in FT-Raman spectra of milk powder samples. PCA and PLS was used with Raman spectroscopic data to quantify the adulteration rate.182 Multiple adulterants, namely ammonium sulphate, dicyandiamide, melamine, and urea present in the milk powder samples were simultaneously detected by using Raman chemical imaging coupled with mixture analysis algorithms. Differences in the Raman spectra of four chemical adulterants allowed researchers to detect and differentiate these compounds. The strongest Raman bands characterizing the relevant chemical compounds were as follows; 973 cm−1 for ammonium sulphate, 212 cm−1 for dicyandiamide, 673 cm−1 for melamine and 1009 cm−1 for urea.183
Adulteration of maple syrup with corn syrup has been investigated using FT-IR, FT-Raman and NIR spectroscopy. Quantitative analyses of adulterated samples were performed with PLS. PCA-LDA, PLS-CVA, PLS-LDA and PCA-CVA methods were also applied for discriminant analysis, but the best results were obtained with PCA-CVA. Characteristic bands mostly attributed to the presence of carbohydrates were used for creating above-mentioned predictive models.184
Adulterated honey samples with various floral origins containing beet and cane inverts were successfully determined using FT-Raman spectroscopy coupled with PLS and PCA combined with CVA and LDA.146
Detection of paraffin in the adulterated rice samples was investigated. Confocal microscope Raman measurements were performed on the surface of rice samples to obtain information about chemical composition. PCA, SIMCA, PLS-DA, KNN and SVM methods were used to differentiate rice samples from different locations and to detect paraffin in the adulterated rice samples. Although the Raman spectra of rice samples comprised of starch, protein and lipid, researchers were also able to detect the presence of paraffin by following the strong Raman bands at 1062, 1132, 1295, 1417, 1440 and 1462 cm−1.185
Methanol and ethanol content of distilled alcoholic beverages was successfully determined with Raman spectroscopy. Quantification of methanol in mixtures was done using the intensities of methanol and ethanol bands located at 1019 and 879 cm−1, respectively. Collected Raman data was normalized by using acetonitrile as an internal standard in the developed method.186 Nguyen and Wu developed a Raman spectroscopic method to quantify low concentrations of methanol in alcohol. PLS regression was applied to the collected Raman spectra where spectral region between 950 and 1200 cm−1 was specifically used to obtain the calibration curves.187
Identification of meat species is of great importance in order to determine the potential adulteration of meat products with cheaper alternatives. Beattie et al., used Raman spectroscopy and multivariate data analysis to classify adipose tissue samples from different origins. PLS-DA and PC-LDA were employed to classify the samples of chicken, beef, lamp and pork species.188 Sowoidnich et al. used shifted excitation Raman difference spectroscopy for the non-invasive differentiation of meat species, namely beef, pork, chicken and turkey. A clear separation was obtained by employing PCA to the collected Raman data.189 Boyaci et al., used Raman spectroscopy in combination with chemometrics to determine the beef adulteration with horsemeat. Employing PCA on collected Raman data enabled researchers to differentiate beef samples spiked with different rates of horsemeat.190 In a similar study reported by the same research group, extracted fat samples were used to differentiate meat species namely, cattle, sheep, pig, fish, poultry, goat and buffalo. Salami products with different formulations prepared by using these meat species were also investigated with Raman spectroscopy.191 Zajac et al. used IR and FT-Raman spectroscopy to analyze the amino acid composition of the samples in order to determine the content of horse meat in its mixture with beef.192
Conclusion
The use of Raman spectroscopy in food analysis is still in its initial stage despite its great potential in almost every field of food science. Raman has many advantages compared to other food analysis methods, and the use of Raman is increasing day by day. Raman spectroscopy can replace traditional food analyses as it does not require labelling and pre-treatment steps. In addition, Raman spectroscopy is a sensitive, reliable, non-destructive and real-time method. Some important issues that need to be addressed for making Raman spectroscopy a more commonly used method are:a. Investigating the applications for Raman spectroscopy in food analysis. Due to some reasons (cost and scarcity of the instrument in food area etc.), the usage of the Raman system in food analysis didn't use to be a common practice. Over the last two decades, however, these limitations have been partially eliminated, and the number of reported studies in this field has increased. Nevertheless, it is still not sufficient and more studies should be conducted. Results of the Raman system should be correlated with standard methods, and new standard methods should be developed using Raman spectroscopy.
According to our detailed research on the literature, some aspects of food analysis still need to be investigated by using Raman spectroscopy. To our knowledge, mineral and toxin analysis in food studies are among these aspects waiting to be dealt with using Raman spectroscopy.
b. Application-driven databases for common analysis. Although some studies have been carried out in this field, there is still no available database related to Raman spectroscopy in food analysis. Complex food matrices and variations in the systems cause difficulties in preparation of databases. In this review, Raman bands obtained in food analyses were summarized in tables (Tables 1S–9S†). We believe that these tables will be beneficial for researchers in this field. However, more systematic experimental work is needed for the preparation of this database. The Raman system's becoming more widespread in food field will help develop a database.
c. Improving analysis efficiency. The complex nature of the food sample reduces efficiency of the analysis. To overcome this difficulty, some easy preprocessing practices could be conducted before Raman measurement. In addition, novel data processing techniques (chemometric methods and artificial neural networks) will be of more help to the user, and processing Raman spectra with these methods will increase the efficiency of the analysis.
d. Better analyser instrument. Most of the Raman systems were developed for the research purposes in laboratories. There is no individual Raman system for specific food analysis. The success of Raman spectroscopy in food analysis will be increased through developing individual Raman systems. Field assays are also very important in food analysis. Portable Raman systems have big potential in this field. Developments in light source and detector technologies will help produce small size portable Raman modules with better performance.
e. Low cost analyser system. High cost of Raman modules obstructs the common usage of the system in this field. Simpler systems, designed specifically for the sample and portable Raman system should be produced with lower instrumentation cost.
Raman spectroscopy in food analysis is receiving a lot of interest from researchers worldwide. We believe that Raman spectroscopy will be one of the most common methods to be used in food analysis in the near future.
Notes and references
- L. M. Reid, C. P. O'Donnell and G. Downey, Trends Food Sci. Technol., 2006, 17, 344–353 CrossRef CAS PubMed.
- C. N. G. Scotter, Trends Food Sci. Technol., 1997, 8, 285–292 CrossRef CAS.
- D. T. Yang and Y. B. Ying, Appl. Spectrosc. Rev., 2011, 46, 539–560 CrossRef PubMed.
- A. Rohman and Y. B. C. Man, Food Rev. Int., 2012, 28, 97–112 CrossRef PubMed.
- R. J. Dijkstra, F. Ariese, C. Gooijer and U. A. T. Brinkman, TrAC, Trends Anal. Chem., 2005, 24, 304–323 CrossRef CAS PubMed.
- R. L. McCreery, in Raman Spectroscopy for Chemical Analysis, Wiley-Interscience, New York, 2000, pp. 251–291 Search PubMed.
- B. J. Marquardt and J. P. Wold, Lebensm.-Wiss. Technol., 2004, 37, 1–8 CrossRef CAS.
- H. W. Wong, S. M. Choi, D. L. Phillips and C. Y. Ma, Food Chem., 2009, 113, 363–370 CrossRef CAS PubMed.
- A. M. Herrero, Food Chem., 2008, 107, 1642–1651 CrossRef CAS PubMed.
- D. Yang and Y. Ying, Appl. Spectrosc. Rev., 2012, 46, 539–560 CrossRef PubMed.
- E. F. Olsen, C. Baustad, B. Egelandsdal, E. O. Rukke and T. Isaksson, Meat Sci., 2010, 85, 1–6 CrossRef CAS PubMed.
- A. M. Herrero, Crit. Rev. Food Sci. Nutr., 2008, 48, 512–523 CrossRef CAS PubMed.
- H. Schulz and M. Baranska, Vib. Spectrosc., 2007, 43, 13–25 CrossRef CAS PubMed.
- R. Tuma, J. Raman Spectrosc., 2005, 36, 307–319 CrossRef CAS PubMed.
- E. G. Ferrer, A. V. Gomez, M. C. Anon and M. C. Puppo, Spectrochim. Acta, Part A, 2011, 79, 278–281 CrossRef CAS PubMed.
- L. Liao, Q. Wang and M. M. Zhao, J. Sci. Food Agric., 2012, 92, 1865–1873 CrossRef CAS PubMed.
- S. Ngarize, A. Adams and N. K. Howell, Food Hydrocolloids, 2004, 18, 49–59 CrossRef CAS.
- T. W. Barrett, W. L. Peticolas and R. M. Robson, Biophys. J., 1978, 23, 349–358 CrossRef CAS.
- Z. H. Chi, X. G. Chen, J. S. W. Holtz and S. A. Asher, Biochemistry, 1998, 37, 2854–2864 CrossRef CAS PubMed.
- C. Y. Huang, G. Balakrishnan and T. G. Spiro, J. Raman Spectrosc., 2006, 37, 277–282 CrossRef CAS PubMed.
- M. Nonaka, E. Lichan and S. Nakai, J. Agric. Food Chem., 1993, 41, 1176–1181 CrossRef CAS.
- N. Howell and E. LiChan, Int. J. Food Sci. Technol., 1996, 31, 439–451 CAS.
- I. Sánchez-González, P. Carmona, P. Moreno, J. Borderias, I. Sanchez-Alonso, A. Rodriguez-Casado and M. Careche, Food Chem., 2008, 106, 56–64 CrossRef PubMed.
- X.-L. Xu, M.-Y. Han, Y. Fei and G.-H. Zhou, Meat Sci., 2011, 87, 159–164 CrossRef CAS PubMed.
- D. T. Berhe, A. J. Lawaetz, S. B. Engelsen, M. S. Hviid and R. Lametsch, Food Control, 2015, 52, 119–125 CrossRef PubMed.
- Z. L. Lim, N. H. Low, B. A. Moffatt and G. R. Gray, Cryobiology, 2013, 66, 156–166 CrossRef CAS PubMed.
- S. Pieters, Y. Vander Heyden, J.-M. Roger, M. D'Hondt, L. Hansen, B. Palagos, B. De Spiegeleer, J.-P. Remon, C. Vervaet and T. De Beer, Eur. J. Pharm. Biopharm., 2013, 85, 263–271 CrossRef CAS PubMed.
- H.-W. Wong, S.-M. Choi, D. L. Phillips and C.-Y. Ma, Food Chem., 2009, 113, 363–370 CrossRef CAS PubMed.
- Z. L. Kang, P. Wang, X. L. Xu, C. Z. Zhu, K. Li and G. H. Zhou, Meat Sci., 2014, 98, 171–177 CrossRef CAS PubMed.
- J.-H. Shao, Y.-F. Zou, X.-L. Xu, J.-Q. Wu and G.-H. Zhou, Food Res. Int., 2011, 44, 2955–2961 CrossRef CAS PubMed.
- G. T. Meng, J. C. K. Chan, D. Rousseau and E. C. Y. Li-Chan, J. Agric. Food Chem., 2005, 53, 845–852 CrossRef CAS PubMed.
- A. S. Sivam, D. Sun-Waterhouse, C. O. Perera and G. I. N. Waterhouse, Food Res. Int., 2013, 50, 574–585 CrossRef CAS PubMed.
- A. V. Gómez, E. G. Ferrer, M. C. Añón and M. C. Puppo, J. Mol. Struct., 2013, 1033, 51–58 CrossRef PubMed.
- N. Perisic, N. K. Afseth, R. Ofstad, S. Hassani and A. Kohler, Food Chem., 2013, 138, 679–686 CrossRef CAS PubMed.
- D. W. Gruenwedel and J. R. Whitaker, Food analysis: principles and techniques, Dekker, New York, 1984 Search PubMed.
- M. R. Almeida, R. S. Alves, L. B. L. R. Nascimbem, R. Stephani, R. J. Poppi and L. F. C. de Oliveira, Anal. Bioanal. Chem., 2010, 397, 2693–2701 CrossRef CAS PubMed.
- I. Delfino, C. Camerlingo, M. Portaccio, B. Della Ventura, L. Mita, D. G. Mita and M. Lepore, Food Chem., 2011, 127, 735–742 CrossRef CAS PubMed.
- K. Ilaslan, I. H. Boyaci and A. Topcu, Food Control, 2015, 48, 56–61 CrossRef CAS PubMed.
- M. Roman, J. C. Dobrowolski, M. Baranska and R. Baranski, J. Nat. Prod., 2011, 74, 1757–1763 CrossRef CAS PubMed.
- A.-S. Jääskeläinen, U. Holopainen-Mantila, T. Tamminen and T. Vuorinen, J. Cereal Sci., 2013, 57, 543–550 CrossRef PubMed.
- L. Scudiero and C. F. Morris, J. Cereal Sci., 2010, 52, 136–142 CrossRef CAS PubMed.
- N. Wellner, D. M. R. Georget, M. L. Parker and V. J. Morris, Starch - Stärke, 2011, 63, 128–138 CrossRef CAS PubMed.
- N. Linlaud, E. Ferrer, M. A. C. Puppo and C. Ferrero, J. Agric. Food Chem., 2010, 59, 713–719 CrossRef PubMed.
- M. S. Mikkelsen, B. M. Jespersen, F. H. Larsen, A. Blennow and S. B. Engelsen, Food Chem., 2013, 136, 130–138 CrossRef CAS PubMed.
- C. K. Chong, J. Xing, D. L. Phillips and H. Corke, J. Agric. Food Chem., 2001, 49, 2702–2708 CrossRef CAS PubMed.
- D. L. Wetzel, Y. C. Shi and U. Schmidt, Vib. Spectrosc., 2010, 53, 173–177 CrossRef CAS PubMed.
- N. Dupuy and J. Laureyns, Carbohydr. Polym., 2002, 49, 83–90 CrossRef CAS.
- L. Passauer, H. Bender and S. Fischer, Carbohydr. Polym., 2010, 82, 809–814 CrossRef CAS PubMed.
- B. Volkert, A. Lehmann, T. Greco and M. H. Nejad, Carbohydr. Polym., 2010, 79, 571–577 CrossRef CAS PubMed.
- E. Pigorsch, Starch - Stärke, 2009, 61, 129–138 CrossRef CAS PubMed.
- S. N. Yuen, S. M. Choi, D. L. Phillips and C. Y. Ma, Food Chem., 2009, 114, 1091–1098 CrossRef CAS PubMed.
- K. C. Schuster, E. Urlaub and J. R. Gapes, J. Microbiol. Methods, 2000, 42, 29–38 CrossRef CAS.
- P. M. Fechner, S. Wartewig, P. Kleinebudde and R. H. H. Neubert, Carbohydr. Res., 2005, 340, 2563–2568 CrossRef CAS PubMed.
- A. Flores-Morales, M. Jimenez-Estrada and R. Mora-Escobedo, Carbohydr. Res., 2012, 87, 61–68 CrossRef CAS PubMed.
- C. Mutungi, L. Passauer, C. Onyango, D. Jaros and H. Rohm, Carbohydr. Res., 2012, 87, 598–606 CrossRef CAS PubMed.
- M. I. U. Islam and T. A. G. Langrish, Food Res. Int., 2010, 43, 46–56 CrossRef CAS PubMed.
- H. Akinosho, S. Hawkins and L. Wicker, Carbohydr. Polym., 2013, 98, 276–281 CrossRef CAS PubMed.
- R. Kizil and J. Irudayaraj, J. Sci. Food Agric., 2007, 87, 1244–1251 CrossRef CAS PubMed.
- M. Łabanowska, A. Wesełucha-Birczyńska, M. Kurdziel and P. Puch, Carbohydr. Polym., 2013, 92, 842–848 CrossRef PubMed.
- J. Huen, C. Weikusat, M. Bayer-Giraldi, I. Weikusat, L. Ringer and K. Losche, J. Cereal Sci., 2014, 60, 555–560 CrossRef CAS PubMed.
- H. Sadeghijorabchi, P. J. Hendra, R. H. Wilson and P. S. Belton, J. Am. Oil Chem. Soc., 1990, 67, 483–486 CrossRef CAS.
- F. L. Silveira, L. Silveira, A. B. Villaverde, M. T. T. Pacheco and C. A. Pasqualucci, Instrum. Sci. Technol., 2010, 38, 107–123 CrossRef CAS PubMed.
- R. M. El-Abassy, P. J. Eravuchira, P. Donfack, B. von der Kammer and A. Materny, Vib. Spectrosc., 2011, 56, 3–8 CrossRef CAS PubMed.
- C. M. McGoverin, A. S. S. Clark, S. E. Holroyd and K. C. Gordon, Anal. Chim. Acta, 2010, 673, 26–32 CrossRef CAS PubMed.
- J. R. Beattie, S. E. J. Bell, C. Borgaard, A. M. Fearon and B. W. Moss, Lipids, 2004, 39, 897–906 CrossRef CAS PubMed.
- O. Abbas, J. A. F. Pierna, R. Codony, C. von Holst and V. Baeten, J. Mol. Struct., 2009, 924–926, 294–300 CrossRef CAS PubMed.
- V. Baeten, P. Hourant, M. T. Morales and R. Aparicio, J. Agric. Food Chem., 1998, 46, 2638–2646 CrossRef CAS.
- H. Yang, J. Irudayaraj and M. M. Paradkar, Food Chem., 2005, 93, 25–32 CrossRef CAS PubMed.
- B. Muik, B. Lendl, A. Molina-Diaz and M. J. Ayora-Canada, Chem. Phys. Lipids, 2005, 134, 173–182 CrossRef CAS PubMed.
- P. Kathirvel, I. V. Ermakov, W. Gellermann, J. Mai and M. P. Richards, Int. J. Food Sci. Technol., 2008, 43, 2095–2099 CrossRef CAS PubMed.
- E. Guzmán, V. Baeten, J. A. F. Pierna and J. A. Garcia-Mesa, Food Control, 2011, 22, 2036–2040 CrossRef PubMed.
- I. Sanchez-Alonso, P. Carmona and M. Careche, Food Chem., 2012, 132, 160–167 CrossRef CAS PubMed.
- Y. Fan, J. Zhou and D. P. Xu, Spectrochim. Acta, Part A, 2014, 129, 143–147 CrossRef PubMed.
- R. Rodríguez-Solana, D. J. Daferera, C. Mitsi, P. Trigas, M. Polissiou and P. A. Tarantilis, Ind. Crops Prod., 2014, 62, 22–33 CrossRef PubMed.
- R. M. El-Abassy, P. Donfack and A. Materny, Food Res. Int., 2010, 43, 694–700 CrossRef CAS PubMed.
- S. Ötleş, Methods of analysis of food components and additives, CRC Press, Boca Raton, 2005 Search PubMed.
- L. Rimai, D. Gill and J. L. Parsons, J. Am. Chem. Soc., 1971, 93, 1353–1357 CrossRef CAS.
- C. Y. Panicker, H. T. Varghese and D. Philip, Spectrochim. Acta, Part A, 2006, 65, 802–804 CrossRef PubMed.
- C. W. Tsai and M. D. Morris, Anal. Chim. Acta, 1975, 76, 193–198 CrossRef CAS.
- C. Cimpoiu, D. Casoni, A. Hosu, V. Miclaus, T. Hodisan and G. Damian, J. Liq. Chromatogr. Relat. Technol., 2005, 28, 2551–2559 CrossRef CAS PubMed.
- M. Kim and P. R. Carey, J. Am. Chem. Soc., 1993, 115, 7015–7016 CrossRef CAS.
- J. R. Beattie, C. Maguire, S. Gilchrist, L. J. Barrett, C. E. Cross, F. Possmayer, M. Ennis, J. S. Elborn, W. J. Curry, J. J. McGarvey and B. C. Schock, FASEB J., 2007, 21, 766–776 CrossRef CAS PubMed.
- H. Yang and J. Irudayaraj, J. Pharm. Pharmacol., 2002, 54, 1247–1255 CrossRef CAS PubMed.
- V. Velusamy, K. Arshak, O. Korostynska, K. Oliwa and C. Adley, Biotechnol. Adv., 2010, 28, 232–254 CrossRef CAS PubMed.
- M. Koopmans, C. H. von Bonsdorff, J. Vinje, D. de Medici and S. Monroe, FEMS Microbiol. Rev., 2002, 26, 187–205 CAS.
- P. Rösch, M. Schmitt, W. Kiefer and J. Popp, J. Mol. Struct., 2003, 661, 363–369 CrossRef PubMed.
- H. Yang and J. Irudayaraj, J. Mol. Struct., 2003, 646, 35–43 CrossRef CAS.
- S. Stockel, S. Meisel, R. Bohme, M. Elschner, P. Rosch and J. Popp, J. Raman Spectrosc., 2009, 40, 1469–1477 CrossRef PubMed.
- K. Maquelin, L. P. Choo-Smith, T. van Vreeswijk, H. P. Endtz, B. Smith, R. Bennett, H. A. Bruining and G. J. Puppels, Anal. Chem., 2000, 72, 12–19 CrossRef CAS.
- M. Harz, P. Rosch, K. D. Peschke, O. Ronneberger, H. Burkhardt and J. Popp, Analyst, 2005, 130, 1543–1550 RSC.
- E. C. Lopez-Diez and R. Goodacre, Anal. Chem., 2004, 76, 585–591 CrossRef CAS PubMed.
- K. Maquelin, C. Kirschner, L. P. Choo-Smith, N. van den Braak, H. P. Endtz, D. Naumann and G. J. Puppels, J. Microbiol. Methods, 2002, 51, 255–271 CrossRef CAS.
- D. Kusic, B. Kampe, P. Rosch and J. Popp, Water Res., 2014, 48, 179–189 CrossRef CAS PubMed.
- S. Meisel, S. Stockel, P. Rosch and J. Popp, Food Microbiol., 2014, 38, 36–43 CrossRef CAS PubMed.
- U. Münchberg, P. Rosch, M. Bauer and J. Popp, Anal. Bioanal. Chem., 2014, 406, 3041–3050 CrossRef PubMed.
- A. Silge, W. Schumacher, P. Rosch, P. A. Da Costa, C. Gerard and J. Popp, Syst. Appl. Microbiol., 2014, 37, 360–367 CrossRef CAS PubMed.
- H. H. Wang, S. J. Ding, G. Y. Wang, X. L. Xu and G. H. Zhou, Int. J. Food Microbiol., 2013, 167, 293–302 CrossRef CAS PubMed.
- K. De Gussem, P. Vandenabeele, A. Verbeken and L. Moens, Spectrochim. Acta, Part A, 2005, 61, 2896–2908 CrossRef PubMed.
- U. Münchberg, L. Wagner, E. T. Spielberg, K. Voigt, P. Rösch and J. Popp, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2013, 1831, 341–349 CrossRef PubMed.
- E. W. Blanch, L. Hecht and L. D. Barron, Methods, 2003, 29, 196–209 CrossRef CAS.
- E. W. Blanch, I. H. McColl, L. Hecht, K. Nielsen and L. D. Barron, Vib. Spectrosc., 2004, 35, 87–92 CrossRef CAS PubMed.
- R. Tuma and G. J. Thomas, Biophys. Chem., 1997, 68, 17–31 CrossRef CAS.
- Z. Q. Wen, S. A. Overman and G. J. Thomas, Biochemistry, 1997, 36, 7810–7820 CrossRef CAS PubMed.
- Z. Q. Wen and G. J. Thomas, Biopolymers, 1998, 45, 247–256 CrossRef CAS.
- D. Dinakarpandian, B. Shenoy, M. PusztaiCarey, B. A. Malcolm and P. R. Carey, Biochemistry, 1997, 36, 4943–4948 CrossRef CAS PubMed.
- H. Peters, Y. Y. Kusov, S. Meyer, A. J. Benie, E. Bauml, M. Wolff, C. Rademacher, T. Peters and V. Gauss-Muller, Biochem. J., 2005, 385, 363–370 CrossRef CAS PubMed.
- P. Carmona, M. Molina and A. Rodriguez-Casado, J. Raman Spectrosc., 2009, 40, 893–897 CrossRef CAS PubMed.
- A. Rodriguez-Casado, M. Molina and P. Carmona, Appl. Spectrosc., 2007, 61, 1219–1224 CrossRef CAS PubMed.
- A. Rodriguez-Casado, M. Molina and P. Carmona, Proteins, 2007, 66, 110–117 CrossRef CAS PubMed.
- G. Font, M. J. Ruiz, M. Fernández and Y. Picó, Electrophoresis, 2008, 29, 2059–2078 CrossRef CAS PubMed.
- N. N. Brandt, A. Y. Chikishev, A. I. Sotnikov, Y. A. Savochkina, I. I. Agapov and A. G. Tonevitsky, J. Mol. Struct., 2005, 735–736, 293–298 CrossRef CAS PubMed.
- P. X. Zhang, Z. Xiaofang, Y. S. C. Andrew and F. Yan, J. Phys.: Conf. Ser., 2006, 28, 7–11 CrossRef CAS.
- S. Bonora, E. Benassi, A. Maris, V. Tugnoli, S. Ottani and M. Di Foggia, J. Mol. Struct., 2013, 1040, 139–148 CrossRef CAS PubMed.
- G. D. Fleming, J. Villagrán and R. Koch, Spectrochim. Acta, Part A, 2013, 114, 120–128 CrossRef CAS PubMed.
- K. J. Leonard and W. R. Bushnell, Fusarium head blight of wheat and barley, APS Press, MN, Saint Paul, 2004 Search PubMed.
- J. E. Dexter, R. M. Clear and K. R. Preston, Cereal Chem., 1996, 73, 695–701 CAS.
- W. L. Casale, J. J. Pestka and L. P. Hart, J. Agric. Food Chem., 1988, 36, 663–668 CrossRef CAS.
- Y. Liu, S. R. Delwiche and Y. Dong, Food Addit. Contam., Part A, 2009, 26, 1396–1401 CrossRef CAS PubMed.
- K. M. Lee, T. J. Herrman and U. Yun, J. Cereal Sci., 2014, 59, 70–78 CrossRef PubMed.
- K.-M. Lee, J. Davis, T. J. Herrman, S. C. Murray and Y. Deng, Food Chem., 2015, 173, 629–639 CrossRef CAS PubMed.
- G. Gupta, A. S. B. Bhaskar, B. K. Tripathi, P. Pandey, M. Boopathi, P. V. L. Rao, B. Singh and R. Vijayaraghavan, Biosens. Bioelectron., 2011, 26, 2534–2540 CrossRef CAS PubMed.
- C. Sproll, W. Ruge, C. Andlauer, R. Godelmann and D. W. Lachenmeier, Food Chem., 2008, 109, 462–469 CrossRef CAS PubMed.
- V. Sortur, J. Yenagi, J. Tonannavar, V. B. Jadhav and M. V. Kulkarni, Spectrochim. Acta, Part A, 2006, 64, 301–307 CrossRef PubMed.
- Y. Q. Huang, C. K. C. Wong, J. S. Zheng, H. Bouwman, R. Barra, B. Wahlström, L. Neretin and M. H. Wong, Environ. Int., 2012, 42, 91–99 CrossRef CAS PubMed.
- J. Dybal, P. Schmidt, J. Baldrian and J. Kratochvíl, Macromolecules, 1998, 31, 6611–6619 CrossRef CAS.
- T. G. Levitskaia, S. I. Sinkov and S. A. Bryan, Vib. Spectrosc., 2007, 44, 316–323 CrossRef CAS PubMed.
- Z. Yu, L. A. Bracero, L. Chen, W. Song, X. Wang and B. Zhao, Spectrochim. Acta, Part A, 2013, 105, 52–56 CrossRef CAS PubMed.
- S. A. Asher, Anal. Chem., 1984, 56, 720–724 CrossRef CAS.
- A. I. Alajtal, H. G. M. Edwards and I. J. Scowen, J. Raman Spectrosc., 2011, 42, 179–185 CrossRef CAS PubMed.
- N. Sundaraganesan, N. Puviarasan and S. Mohan, Talanta, 2001, 54, 233–241 CrossRef CAS.
- I. V. Ermakov, M. R. Ermakova and W. Gellermann, Proc. Soc. Photo-Opt. Instrum. Eng., 2006, 6078, 7835 CrossRef PubMed.
- E. Miranda-Bermudez, N. Belai, B. P. Harp, B. J. Yakes and J. N. Barrows, Food Addit. Contam., Part A, 2012, 29, 38–42 CrossRef CAS PubMed.
- M. Snehalatha, C. Ravikumar, N. Sekar, V. S. Jayakumar and I. H. Joe, J. Raman Spectrosc., 2008, 39, 928–936 CrossRef CAS PubMed.
- N. Peica, I. Pavel, S. C. Pinzaru, V. K. Rastogi and W. Kiefer, J. Raman Spectrosc., 2005, 36, 657–666 CrossRef CAS PubMed.
- C. S. Mangolim, C. Moriwaki, A. C. Nogueira, F. Sato, M. L. Baesso, A. M. Neto and G. Matioli, Food Chem., 2014, 153, 361–370 CrossRef CAS PubMed.
- K. Zborowski, R. Grybos and L. M. Proniewicz, Vib. Spectrosc., 2005, 37, 233–236 CrossRef CAS PubMed.
- N. Peica, C. Lehene, N. Leopold, S. Schlucker and W. Kiefer, Spectrochim. Acta, Part A, 2007, 66, 604–615 CrossRef CAS PubMed.
- N. Peica, J. Raman Spectrosc., 2009, 40, 2144–2154 CrossRef CAS PubMed.
- T. Jiang, R. James, S. G. Kumbar and C. T. Laurencin, in Natural and Synthetic Biomedical Polymers, 2014, pp. 91–113 Search PubMed.
- A. Zajac, J. Hanuza, M. Wandas and L. Dyminska, Spectrochim. Acta, Part A, 2015, 134, 114–120 CrossRef CAS PubMed.
- K. Zhang, J. Helm, D. Peschel, M. Gruner, T. Groth and S. Fischer, Polymer, 2010, 51, 4698–4705 CrossRef CAS PubMed.
- M. C. Sarraguca, T. De Beer, C. Vervaet, J. P. Remon and J. A. Lopes, Talanta, 2010, 83, 130–138 CrossRef CAS PubMed.
- R. M. El-Abassy, P. Donfack and A. Materny, Food Chem., 2011, 126, 1443–1448 CrossRef CAS PubMed.
- R. Goodacre, B. S. Radovic and E. Anklam, Appl. Spectrosc., 2002, 56, 521–527 CrossRef CAS.
- M. Holse, F. H. Larsen, A. Hansen and S. B. Engelsen, Food Res. Int., 2011, 44, 373–384 CrossRef CAS PubMed.
- M. M. Paradkar and J. Irudayaraj, Food Chem., 2002, 76, 231–239 CrossRef CAS.
- B. Özbalci, I. H. Boyaci, A. Topcu, C. Kadılar and U. Tamer, Food Chem., 2013, 136, 1444–1452 CrossRef PubMed.
- F. Corvucci, L. Nobili, D. Melucci and F.-V. Grillenzoni, Food Chem., 2015, 169, 297–304 CrossRef CAS PubMed.
- A. Keidel, D. von Stetten, C. Rodrigues, C. MaÃÅguas and P. Hildebrandt, J. Agric. Food Chem., 2010, 58, 11187–11192 CrossRef CAS PubMed.
- A. B. Rubayiza and M. Meurens, J. Agric. Food Chem., 2005, 53, 4654–4659 CrossRef CAS PubMed.
- R. Korifi, Y. Le Dreau, J. Molinet, J. Artaud and N. Dupuy, J. Raman Spectrosc., 2011, 42, 1540–1547 CrossRef CAS PubMed.
- E. Guzmán, V. Baeten, J. A. F. Pierna and J. A. Garcia-Mesa, Talanta, 2012, 93, 94–98 CrossRef PubMed.
- I. Gouvinhas, N. Machado, T. Carvalho, J. M. M. M. de Almeida and A. I. R. N. A. Barros, Talanta, 2015, 132, 829–835 CrossRef CAS PubMed.
- H. M. Velioğlu, H. T. Temiz and I. H. Boyaci, Food Chem., 2015, 172, 283–290 CrossRef PubMed.
- H. Mohamadi Monavar, N. K. Afseth, J. Lozano, R. Alimardani, M. Omid and J. P. Wold, Talanta, 2013, 111, 98–104 CrossRef CAS PubMed.
- K. Czamara, K. Majzner, M. Z. Pacia, K. Kochan, A. Kaczor and M. Baranska, J. Raman Spectrosc., 2015, 46, 4–20 CrossRef CAS PubMed.
- S. Sivakesava, J. Irudayaraj and A. Demirci, J. Ind. Microbiol. Biotechnol., 2001, 26, 185–190 CrossRef CAS PubMed.
- S. Sivakesava, J. Irudayaraj and D. Ali, Process Biochem., 2001, 37, 371–378 CrossRef CAS.
- R. S. Uysal, E. A. Soykut, I. H. Boyaci and A. Topcu, Food Chem., 2013, 141, 4333–4343 CrossRef CAS PubMed.
- Q. Y. Wang, Z. G. Li, Z. H. Ma and L. Q. Liang, Sens. Actuators, B, 2014, 202, 426–432 CrossRef CAS PubMed.
- R. Rodriguez, S. Vargas, M. Estevez, F. Quintanilla, A. Trejo-Lopez and A. R. Hernández-Martínez, Vib. Spectrosc., 2013, 68, 133–140 CrossRef CAS PubMed.
- H. Lee, B.-K. Cho, M. S. Kim, W.-H. Lee, J. Tewari, H. Bae, S.-I. Sohn and H.-Y. Chi, Sens. Actuators, B, 2013, 185, 694–700 CrossRef CAS PubMed.
- J. Trebolazabala, M. Maguregui, H. Morillas, A. de Diego and J. M. Madariaga, Spectrochim. Acta, Part A, 2013, 105, 391–399 CrossRef CAS PubMed.
- J. Qin, K. Chao and M. S. Kim, J. Food Eng., 2011, 107, 277–288 CrossRef CAS PubMed.
- A. G. Gonzalvez, D. Martin, K. Slowing and A. Gonzalez Ureña, Food Struct., 2014, 2, 61–65 CrossRef PubMed.
- H. L. T. Lee, P. Boccazzi, N. Gorret, R. J. Ram and A. J. Sinskey, Vib. Spectrosc., 2004, 35, 131–137 CrossRef CAS PubMed.
- Y. Numata, Y. Shinohara, T. Kitayama and H. Tanaka, Process Biochem., 2013, 48, 569–574 CrossRef CAS PubMed.
- A. A. Argyri, R. M. Jarvis, D. Wedge, Y. Xu, E. Z. Panagou, R. Goodacre and G.-J. E. Nychas, Food Control, 2013, 29, 461–470 CrossRef CAS PubMed.
- P. J. Li, B. H. Kong, Q. Chen, D. M. Zheng and N. Liu, Meat Sci., 2013, 93, 67–72 CrossRef CAS PubMed.
- S. K. Oh, S. J. Yoo, D. H. Jeong and J. M. Lee, Bioresour. Technol., 2013, 142, 131–137 CrossRef CAS PubMed.
- O. Abbas, P. Dardenne and V. Baeten, in Chemical Analysis of Food: Techniques and Applications, Academic Press, Boston, 2012, pp. 59–89 Search PubMed.
- M. Q. Zou, X. F. Zhang, X. H. Qi, H. L. Ma, Y. Dong, C. W. Liu, X. Guo and H. Wang, J. Agric. Food Chem., 2009, 57, 6001–6006 CrossRef CAS PubMed.
- X. F. Zhang, M. Q. Zou, X. H. Qi, F. Liu, C. Zhang and F. Yin, J. Raman Spectrosc., 2011, 42, 1784–1788 CrossRef CAS PubMed.
- X. F. Zhang, X. H. Qi, M. Q. Zou and F. Liu, Anal. Lett., 2011, 44, 2209–2220 CrossRef CAS PubMed.
- E. C. Lopez-Diez, G. Bianchi and R. Goodacre, J. Agric. Food Chem., 2003, 51, 6145–6150 CrossRef CAS PubMed.
- R. M. El-Abassy, P. Donfack and A. Materny, J. Raman Spectrosc., 2009, 40, 1284–1289 CrossRef CAS PubMed.
- H. Yang and J. Irudayaraj, J. Am. Oil Chem. Soc., 2001, 78, 889–895 CrossRef CAS PubMed.
- V. Baeten and R. Aparicio, Biotechnol., Agron., Soc. Environ., 2000, 4, 196–203 CAS.
- R. S. Uysal, I. H. Boyaci, H. E. Genis and U. Tamer, Food Chem., 2013, 141, 4397–4403 CrossRef CAS PubMed.
- Y. Cheng, Y. Y. Dong, J. H. Wu, X. R. Yang, H. Bai, H. Y. Zheng, D. M. Ren, Y. D. Zou and M. Li, J. Food Compos. Anal., 2010, 23, 199–202 CrossRef CAS PubMed.
- S. Okazaki, M. Hiramatsu, K. Gonmori, O. Suzuki and A. T. Tu, Forensic Toxicol., 2009, 27, 94–97 CrossRef CAS PubMed.
- G. P. S. Smith, K. C. Gordon and S. E. Holroyd, Vib. Spectrosc., 2013, 67, 87–91 CrossRef CAS PubMed.
- J. Qin, K. Chao and M. S. Kim, Food Chem., 2013, 138, 998–1007 CrossRef CAS PubMed.
- M. M. Paradkar, S. Sakhamuri and J. Irudayaraj, J. Food Sci., 2002, 67, 2009–2015 CrossRef CAS PubMed.
- X. Feng, Q. Zhang, P. Cong and Z. Zhu, Talanta, 2013, 115, 548–555 CrossRef CAS PubMed.
- I. H. Boyaci, H. E. Genis, B. Guven, U. Tamer and N. Alper, J. Raman Spectrosc., 2012, 43, 1171–1176 CrossRef CAS PubMed.
- D. Nguyen and E. Wu, Spectroscopy, 2011, 26, 95 Search PubMed.
- J. R. Beattie, S. J. Bell, C. Borggaard, A. Fearon and B. Moss, Lipids, 2007, 42, 679–685 CrossRef CAS PubMed.
- K. Sowoidnich and H.-D. Kronfeldt, Appl. Phys. B: Lasers Opt., 2012, 108, 975–982 CrossRef CAS PubMed.
- I. H. Boyaci, H. T. Temiz, R. S. Uysal, H. M. Velioğlu, R. J. Yadegari and M. M. Rishkan, Food Chem., 2014, 148, 37–41 CrossRef CAS PubMed.
- İ. Boyaci, R. Uysal, T. Temiz, E. Shendi, R. Yadegari, M. Rishkan, H. Velioglu, U. Tamer, D. Ozay and H. Vural, Eur. Food Res. Technol., 2014, 238, 845–852 CrossRef.
- A. Zając, J. Hanuza and L. Dymińska, Food Chem., 2014, 156, 333–338 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: ESI data including the tables displaying the Raman bands of food components (proteins, carbohydrates, lipids, and vitamins), microorganisms and viruses, toxins and chemicals, food additives, raw materials and food adulterant is available. See DOI: 10.1039/c4ra12463d |
| This journal is © The Royal Society of Chemistry 2015 |