Optimization and partial purification of a high-activity lipase synthesized by a newly isolated Acinetobacter from offshore waters of the Caspian Sea under solid-state fermentation
Samira Moradia,
Seyed Hadi Razavi*a,
Seyyed Mohammad Mousavib and
Seyed Mohammad Taghi Gharibzahedia
aBioprocess Engineering Laboratory (BPEL), Department of Food Science, Engineering & Technology, Faculty of Agricultural Engineering and Technology, University of Tehran, P.O. Box 4111, Karaj 31587-77871, Iran. E-mail: srazavi@ut.ac.ir; Fax: +98 26 3224 9453; Tel: +98 26 3224 8804
bBiotechnology Group, Chemical Engineering Department, Faculty of Engineering, Tarbiat Modares University, Tehran, Iran
First published on 14th January 2015
Abstract
A new aerobic mesophilic bacterium was isolated from the southern coastal waters of the Caspian Sea which substantially produced an extracellular lipase in solid-state fermentation using milled coriander seeds (MCS) as support substrate. This bacterium was identified as a strain of genus Acinetobacter based on morphological and biochemical characterization and 16S rRNA gene sequence. The various medium components and culture parameters to achieve a more cost effective and economically viable bioprocess were screened and optimized using the Plackett–Burman and central composite designs. The highest lipase activity (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480.2 U g−1) was achieved at optimum levels of predominant factors of MCS/yeast extract (4.0 w/w), olive oil concentration (30 g L−1), moisture content (65.0%), and agitation rate (180.0 rpm). The enzyme with molecular weight of 46 kDa was purified 26.9-fold to homogeneity by ammonium sulfate precipitation and phenyl-Sepharose hydrophobic interaction chromatography. The functional groups of the lipase were also assigned using Fourier transform-infrared spectroscopy.
480.2 U g−1) was achieved at optimum levels of predominant factors of MCS/yeast extract (4.0 w/w), olive oil concentration (30 g L−1), moisture content (65.0%), and agitation rate (180.0 rpm). The enzyme with molecular weight of 46 kDa was purified 26.9-fold to homogeneity by ammonium sulfate precipitation and phenyl-Sepharose hydrophobic interaction chromatography. The functional groups of the lipase were also assigned using Fourier transform-infrared spectroscopy.
1 Introduction
Lipase (E.C. 3.1.1.3) enzymes are carboxyl esterases that catalyze the hydrolysis of acylglycerols composed of long-chain fatty acids with more than 10 carbon atoms.1 These enzymes have attracted scientific and commercial attention over the past few years, because they are industrially considered to be the most important enzyme groups after proteases and amylases.2 Although lipases are produced by animals, plants and microorganisms, they are mainly synthesized by biotechnological methods using bacteria and fungi.3 The microbial lipases are extracellular metabolites with high potential to catalyze a broad range of reactions in the different aqueous and non-aqueous phases.4 These biocatalysts have interesting characteristics such as high activity under mild conditions, suitable stability in organic solvents, high substrate specificity, and regio- and enantio-selectivity.5 It was demonstrated that the genera Acinetobacter, Pseudomonas and Burkholderia among other microbial strains had the unique activities at a wide range of pH and temperature, and have shown the excellent properties of chemo-, regio-, and enantio-selectivity that made them the interest biocatalysts to use by most organic chemists and pharmacologists.3Although submerged liquid fermentations (SLFs) nowadays are the most important of commercial bioprocesses, the lipase production using solid state fermentations (SSFs) due to the significant reduction of final product cost by decreasing downstream processes is recently considered.6 Microbes in the SSF process are grown on a porous solid substrate in the absence of free water. Large number of agricultural/food residues have been used as suitable substrates for SSF. The growth of cells is provided by the water and nutrient absorbed on the surface of the solid support and within the support matrix.7
In the present study, the novel extracellular lipases from various natural sources such as Caspian Sea water (CSW), extract olive (EO), linseed cake (LC), soy bean cake (SBC), cotton seed cake (CSC), fat milk (FM), olive oil pressing waste (OPW) and dairy industrial effluent (DIE) were isolated and partially purified. The cheap carbon substrates including milled seeds of grape, pomegranate and coriander were also analyzed to achieve the best conditions for batch fermentation process. Response surface methodology (RSM) was then applied to determine the optimal culture media for lipase SSF-production with the highest activity using the best bacteria isolate. RSM development for this bioprocess can result in improved product yields, reduced process variability, and closer conformance of the output responses to nominal and target requirements, thus reducing the development time and the overall costs.8,9
2 Materials and methods
Chemicals and raw materials
p-Nitro phenol (p-NP) and p-nitro phenyl palmitate (p-NPP) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). Rhodamine B, nutrient broth (NB), peptone, yeast extract, iso-propanol, Triton X-100, Tris–HCl, gum arabic and agar were provided by Merck Chemical Co. (Darmstadt, Germany). Coriander, grape and pomegranate seeds and olive oil were purchased from a local market in Tehran (Iran). The contents of protein, lipid and dry matter for the milled seeds of coriander, grape and pomegranate were 15.99, 12.90, and 12.35%, 15.50, 24.85 and 20.05%, and 96.1, 93.2 and 93.4%, respectively. All other chemicals were of analytical grade.Isolation and screening of lipolytic bacteria
Various natural sources including CSW, EO, LC, SBC, CSC, FM, OPW and DIE were used to isolate a bacterial strain with the highest lipolytic activity. 1.0 g each sample to enrich was cultured into 250 mL-Erlenmeyer flasks containing 50 mL of broth (NB, 8 g L−1 (w/v); NaCl, 3 g L−1 (w/v); and emulsified olive oil (EOO), 25 g L−1 (w/v)) at pH 6.5. The flasks were incubated at 37 °C with a constant shaking rate of 160 rpm for 24 h. Initial qualitative screening for lipase producing isolate was conducted by serially spreading the diluted samples on a specific medium containing 10 g L−1 extract yeast, 3 g L−1 NaCl, 25 g L−1 EOO, 0.01 g L−1 rhodamine B and 20 g L−1 agar at pH 7.0. Incubation temperature for the plates was chosen in range of 20–45 °C. The formation of orange fluorescent halos around colonies under ultraviolet light (350 nm) indicates lipase positive isolates. Bacterial colony with the largest halo were isolated by repeated pure culture technique and stored for the further studies according to the described method by Sengun et al.10 with some modifications. Briefly, after the purification and isolation steps, the number of isolated colonies using sampler tip were collected and inoculated into the test tubes containing 10 g L−1 yeast extract, 3 g L−1 sodium chloride and 25 g L−1 EOO (pH = 6.5). The test tubes were then incubated at 37 °C for 24 h in order to achieve the favorable growth and suitable turbidity. In the next step, 1000 μL of microbial suspension was transferred to a 1.5 mL pre-sterilized microtube and centrifuged at 4000 rpm for 15 min. After the discarding supernatant, 500 μL of the suspension of the isolated microbe and 500 μL of pre-sterilized liquid culture containing 30% glycerol added to the microbial sediment of microtube bottom. Glycerol concentration in the final solution should be 15%.Selection of the best lipase-producing bacteria isolate
One loopful from orange halo colonies of fresh cultures was inoculated into a medium containing of 10 g L−1 extract yeast, 3 g L−1 NaCl and 25 g L−1 EOO at pH 7.0, and then incubated for 24 h at 37 °C under a shaking rate of 160 rpm. Then, the cultures were centrifuged twice at 6000 rpm for 30 min (Hettich Centrifuge, D-78532 model, Tuttlingen, Germany), filtered through 0.2 μm filters to remove the cell mass and other solids. The obtained supernatant was used in lipase activity (LA) assay.Morphological, biochemical and molecular identification of selected isolate
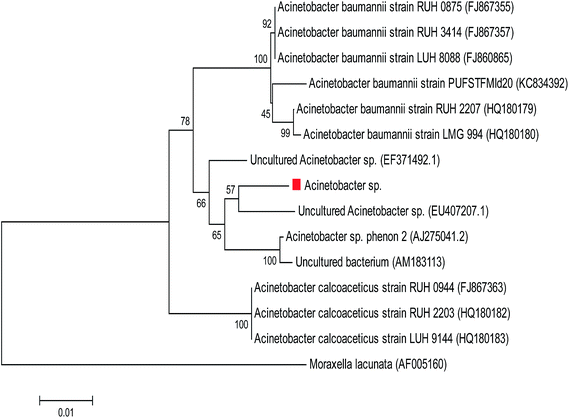
The selected bacterium was identified by morphological and biochemical properties according to Bergey's Manual of Determinative Bacteriology.11 The identification was further confirmed by the 16S rRNA gene sequencing method. The genomic DNA of isolate was extracted as previously described by Ausubel et al.12 The DNA was then amplified by PCR using the following universal 16S ribosomal RNA (rRNA) gene primers, 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The amplification of 16S rRNA gene was conducted in BioRad PCR cycler (USA). PCR was carried out by subjecting a reaction mixture to initial denaturation at 94 °C for 2 min, followed by 35 cycles of 94 °C for 45 s, 55 °C for 1 min, 72 °C for 1.5 min and a final extension step at 72 °C for 15 min. The 16S rRNA gene sequence was compared with sequences available in the nucleotide database using the BLAST algorithm at the NCBI server. Phylogenetic tree was constructed by the neighbor joining method using molecular evolutionary genetics analysis (MEGA) software version 4.11Solid-state fermentation process
The batch experiments for lipase production were carried out in 500 mL-Erlenmeyer flasks based on the milled seeds of coriander, grape and pomegranate. These solid substrates were prepared by washing with distilled water and next air-drying. The powders were sieved and divided to two major fractions based on the particle diameter of 1–2 mm (Tyler Standard Sieve Series of 9–16) and 0.5–1 mm (Tyler Standard Sieve Series of 16–32). Owing to the low accessibility of bacterial cells to nutrients and inappropriate aeration in the cultures with substrate particles size more than 1 mm, the fraction with particle diameter of 0.5–1 mm was selected for use in the fermentation process because of the better cell growth and higher enzyme production.A stable oil-in-water emulsion with 20% w/w gum arabic was prepared to use olive oil (15% w/w) in the culture medium. The coarse emulsion was produced by Ultra-Turrax (IKA T25 Digital, Germany) at 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2.5 min and, further emulsification was performed using a 20 kHz-ultrasounic homogenizer (UP200S, Hielscher Ultrasonics GmbH, Teltow, Germany) equipped to a water bath (45 °C) at a total nominal output power of 45 W for 3 min.8 Each sterilized flask was inoculated with the constant amount of the active isolate (10%) and then incubated at 37 °C for 72 h. Activation of the stored bacterium by placing at ambient laboratory conditions (in a sterile environment under the hood) was started. When it was a little freezing outside, amount of the liquid into microtube using sampler tip was transferred to the culture medium containing 10 g L−1 yeast extract, 3 g L−1 sodium chloride and 25 g L−1 EOO (pH = 6.5) and incubated at 37 °C for 18 h (Fig. 1). According to the RSM design, the used ranges for ratio of solid substrate to yeast extract, olive oil, moisture content (MC) and agitation rate were 3.33 to 6.01 (w/w), 7.5 to 37.5 g L−1, 57.5–87.5% (w.b.), and 135 to 195 rpm, respectively.
000 rpm for 2.5 min and, further emulsification was performed using a 20 kHz-ultrasounic homogenizer (UP200S, Hielscher Ultrasonics GmbH, Teltow, Germany) equipped to a water bath (45 °C) at a total nominal output power of 45 W for 3 min.8 Each sterilized flask was inoculated with the constant amount of the active isolate (10%) and then incubated at 37 °C for 72 h. Activation of the stored bacterium by placing at ambient laboratory conditions (in a sterile environment under the hood) was started. When it was a little freezing outside, amount of the liquid into microtube using sampler tip was transferred to the culture medium containing 10 g L−1 yeast extract, 3 g L−1 sodium chloride and 25 g L−1 EOO (pH = 6.5) and incubated at 37 °C for 18 h (Fig. 1). According to the RSM design, the used ranges for ratio of solid substrate to yeast extract, olive oil, moisture content (MC) and agitation rate were 3.33 to 6.01 (w/w), 7.5 to 37.5 g L−1, 57.5–87.5% (w.b.), and 135 to 195 rpm, respectively.
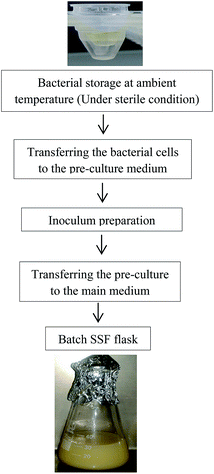 | ||
| Fig. 1 The bacterial activation, inoculation steps and preparation of batch SSF flask for the lipase production. | ||
Lipolytic activity determination
The LA was assayed by measuring the absorbance increase at 410 nm using an UV-visible spectrophotometer (CE2502, BioQuest & BioAquarius Series, Cecil Instr. Inc., Cambridge, UK). This increase was due to the p-NPP hydrolysis and p-NP release at pH 8.0 (37 °C) after 30 min of reaction time. To initialize the reaction, 0.1 mL of the resulted supernatant was added to 0.9 mL of substrate solution containing 3 mg p-NPP dissolved in 1 mL isopropanol diluted in 9 mL of 50 mM Tris–HCl (pH 8.0) containing 40 mg of Triton X-100 and 10 mg of gum arabic.13 One unit (U) of lipase was defined as the amount of enzyme that releases 1 μmol p-NP per min under the assay conditions. The calibration curve with p-NP as standard was prepared.Statistical analysis
| A run | Independent variables | LAd (U g−1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yeast extract (g L−1) | Peptone (g L−1) | Particle size (mm) | MCSa (g L−1) | MPCb (g L−1) | MGCc (g L−1) | MgCl2 (mM) | Agitation rate (rpm) | MC (%) | Molasses (g L−1) | Olive oil (g L−1) | ||
| a MCS: milled coriander seeds.b MPC: milled pomegranate seeds.c MGC: milled grape seeds.d The activity was measured after 24 h incubation. | ||||||||||||
| 1 | 15 | 15 | 0.5 | 40 | 40 | 40 | 20 | 130 | 50 | 15 | 10 | 5452.5 |
| 2 | 5 | 15 | 2 | 20 | 40 | 40 | 40 | 130 | 50 | 5 | 30 | 5234.0 |
| 3 | 15 | 5 | 2 | 40 | 20 | 40 | 40 | 180 | 50 | 5 | 10 | 6543.0 |
| 4 | 5 | 15 | 0.5 | 40 | 40 | 20 | 40 | 180 | 80 | 5 | 10 | 6787.0 |
| 5 | 5 | 5 | 2 | 20 | 40 | 40 | 20 | 180 | 80 | 15 | 10 | 5456.0 |
| 6 | 5 | 5 | 0.5 | 40 | 20 | 40 | 40 | 130 | 80 | 15 | 30 | 6634.0 |
| 7 | 15 | 5 | 0.5 | 20 | 40 | 20 | 40 | 180 | 50 | 15 | 30 | 7431.0 |
| 8 | 15 | 15 | 0.5 | 20 | 20 | 40 | 20 | 180 | 80 | 5 | 30 | 7518.0 |
| 9 | 15 | 15 | 2 | 20 | 20 | 20 | 40 | 130 | 80 | 15 | 10 | 6300.0 |
| 10 | 5 | 15 | 2 | 40 | 20 | 20 | 20 | 180 | 50 | 15 | 30 | 8478.7 |
| 11 | 15 | 5 | 2 | 40 | 40 | 20 | 20 | 130 | 80 | 5 | 30 | 6845.0 |
| 12 | 5 | 5 | 1.25 | 20 | 20 | 20 | 20 | 130 | 50 | 5 | 10 | 5294.0 |
| 13 | 10 | 10 | 1.25 | 30 | 30 | 30 | 30 | 155 | 65 | 10 | 20 | 7638.0 |
| 14 | 10 | 10 | 1.25 | 30 | 30 | 30 | 30 | 155 | 65 | 10 | 20 | 7605.5 |
| Run | Independent variables | Response variable (LA, U g−1) | ||||
|---|---|---|---|---|---|---|
| MCS/yeast extract (w/w) | Olive oil (g L−1) | MC (%) | Agitation rate (rpm) | Experimental valuea | Predicted value | |
| a Mean ± standard deviation (n = 3). | ||||||
| 1 | 4.00 | 15 | 65 | 150 | 7887.5 ± 126 | 9239 |
| 2 | 5.34 | 15 | 65 | 150 | 6510 ± 45 | 6221 |
| 3 | 4.00 | 30 | 65 | 150 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 577 ± 79 577 ± 79 |
9996 |
| 4 | 5.34 | 30 | 65 | 150 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 412 ± 122 412 ± 122 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 323 323 |
| 5 | 4.00 | 15 | 80 | 150 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 792 ± 412 792 ± 412 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 148 148 |
| 6 | 5.34 | 15 | 80 | 150 | 5738 ± 321 | 6546 |
| 7 | 4.00 | 30 | 80 | 150 | 5955 ± 152 | 6556 |
| 8 | 5.34 | 30 | 80 | 150 | 3135 ± 27 | 4298 |
| 9 | 4.00 | 15 | 65 | 180 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 832 ± 119 832 ± 119 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 162 162 |
| 10 | 5.34 | 15 | 65 | 180 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 725 ± 349 725 ± 349 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 102 102 |
| 11 | 4.00 | 30 | 65 | 180 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 310 ± 487 310 ± 487 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480 480 |
| 12 | 5.34 | 30 | 65 | 180 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 627 ± 29 627 ± 29 |
16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 764 764 |
| 13 | 4.00 | 15 | 80 | 180 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027 ± 245 027 ± 245 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 094 094 |
| 14 | 5.34 | 15 | 80 | 180 | 5376 ± 216 | 6450 |
| 15 | 4.00 | 30 | 80 | 180 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 ± 450 280 ± 450 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 062 062 |
| 16 | 5.34 | 30 | 80 | 180 | 5135 ± 32 | 3762 |
| 17 | 3.33 | 22.5 | 72.5 | 165 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 187 ± 161 187 ± 161 |
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 883 883 |
| 18 | 6.01 | 22.5 | 72.5 | 165 | 5736 ± 49 | 4566 |
| 19 | 4.67 | 7.5 | 72.5 | 165 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 497 ± 146 497 ± 146 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 693 693 |
| 20 | 4.67 | 37.5 | 72.5 | 165 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 433 ± 87 433 ± 87 |
9763 |
| 21 | 4.67 | 22.5 | 57.5 | 165 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 632 ± 246 632 ± 246 |
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 663 663 |
| 22 | 4.67 | 22.5 | 87.5 | 165 | 8075 ± 87 | 7570 |
| 23 | 4.67 | 22.5 | 72.5 | 135 | 8713 ± 341 | 7287 |
| 24 | 4.67 | 22.5 | 72.5 | 195 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 723 ± 246 723 ± 246 |
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 675 675 |
| 25 | 4.67 | 22.5 | 72.5 | 165 | 3142 ± 21 | 3387 |
| 26 | 4.67 | 22.5 | 72.5 | 165 | 4651 ± 444 | 3387 |
| 27 | 4.67 | 22.5 | 72.5 | 165 | 3362 ± 12 | 3387 |
| 28 | 4.67 | 22.5 | 72.5 | 165 | 4360 ± 32 | 3387 |
| 29 | 4.67 | 22.5 | 72.5 | 165 | 3271 ± 145 | 3387 |
| 30 | 4.67 | 22.5 | 72.5 | 165 | 3387 ± 64 | 3387 |
| 31 | 4.67 | 22.5 | 72.5 | 165 | 1536 ± 316 | 3387 |
Both linear and quadratic effects of the four variables under study, as well as their interactions, on the dependent variable namely LA (U g−1, Y) were calculated. Their significance was evaluated by analysis of variance (ANOVA). The experimental design results were fitted by a second-order polynomial equation in order to correlate the response to the independent variables. The general equation to predict the optimal point was explained as follows (eqn (1)):
 | (1) |
Optimal conditions for the constructed model solution depended on combination of the independent process variables were obtained through the predictive equation of RSM and the LA. Three additional confirmation experiments were carried out to verify the accuracy of statistical experimental design. Finally, the experimental and predicted values were compared in order to determine the model validity.
The obtained data for LAs produced by the screened bacterial isolates and LA of the Acinetobacter isolated from CSW as a function of different metal ions were subjected to analysis of variance (ANOVA) using SPSS 13 software (SPSS Inc., Chicago, Illinois, USA). The means were compared using the Duncan's multiple ranges test at a significant level of P < 0.05.
Partial purification of the produced lipase
The calculated amount of solid ammonium sulphate was added to cell free supernatant with constant stirring at 4 °C to give a concentration of 80% (w/v) saturation. The precipitate was allowed to settle overnight, harvested by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 min and resuspended in minimum volume of 25 mM sodium phosphate buffer (pH 7.2). This enzyme solution was subjected to dialysis for 24 h at 4 °C against the same buffer, with three intermittent changes of the buffer. The filtrate was further concentrated using centricon centrifugal filters (30 kDa cutoff; Millipore, Billerica, MA) at 800 × g for 1 h at 4 °C. LA and protein content (by the method of Lowry) were determined for both the dialyzed and the concentrated filtrate sample.14
000 × g for 30 min and resuspended in minimum volume of 25 mM sodium phosphate buffer (pH 7.2). This enzyme solution was subjected to dialysis for 24 h at 4 °C against the same buffer, with three intermittent changes of the buffer. The filtrate was further concentrated using centricon centrifugal filters (30 kDa cutoff; Millipore, Billerica, MA) at 800 × g for 1 h at 4 °C. LA and protein content (by the method of Lowry) were determined for both the dialyzed and the concentrated filtrate sample.14
A hydrophobic interaction chromatography (HIC) column of phenyl Sepharose™ (1.5 × 6 cm) pre-equilibrated with the solution containing 0.6 M ammonium sulphate dissolved in 25 mM phosphate buffer (pH 7.2) was used to assay purity of the concentrated lipase solution. This solution was injected to elute the bound enzyme with a flow rate of 1 mL min−1 through negative linear gradient.
Molecular weight determination
The procedure of Laemmli et al.15 was applied to perform the electrophoresis of sodium dodecyl sulphate-polyacrylamide gel (SDS-PAGE on a 5% polyacrylamide stacking gel and a 12% polyacrylamide-resolving gel). A protein marker in range of 14.3 to 94.7 kDa as a standard marker was used to evaluate molecular weight. Protein bands were visualized by silver staining.Fourier transform-infrared spectroscopy (FT-IR)
An infrared absorption spectrum between 500–2000 cm−1 was recorded using a Fourier transform infrared spectroscopy (FT-IR, Model 1725X, Perkin Elmer, Norwalk, CT, USA) with the specimen prepared as a potassium bromide (KBr) pellet in order to determine the functional groups present in the purified lipase. The samples made in pellet form had a thickness of 1 mm and a diameter of 13 mm.3 Results
Preliminary study
The objectives of preliminary experiments were to (I) select the most ideal natural source among CSW, EO, LC, SBC, CSC, FM, OPW and DIE to isolate the lipase-producing bacterium, (II) identify and characterize the best bacterial isolate, (III) determine the most suitable trace element and solid substrate to increase lipase production with the highest activity and then (IV) found the best culture formulation and operation conditions among the various variables.Twelve bacteria were isolated from eight sources by culturing in NB enriched with olive oil. The isolates were screened based on their lipolytic potential using rhodamine B-agar plate. The formation of orange fluorescent halos around colonies ultraviolet light (350 nm) indicates lipase positive isolate. Among the screened isolates, the CSW isolate due to the highest LA (3124 U mL−1) was selected for the further studies (Fig. 2a). The cells of this bacterial isolate were Gram-negative, aerobic, short rod- and coccoid-like, non-motile, non-endospore-forming, non-acid fast, oxidase-negative, and catalase-positive. The selected isolate was positive for H2S and indole production and urea hydrolysis, but they could not reduce nitrates to nitrogen. The biochemical tests of Voges–Proskauer and citrate utilization were negative and positive, respectively. This bacterial isolate was able to utilize the sole carbon sources of D-glucose, D-mannose, D-tagatose, L-rhamnose, lactose, maltose, sucrose, trehalose, gentiobiose, melezitose and melibiose. However, it was unable to utilize inulin, inositol, D-adonitol, D-arabitol, L-arabinose, arbutin, cellobiose, raffinose and cellobiose. Moreover, optimum range for growth temperature, salinity (tolerance level to NaCl) and pH for the selected isolate were 15–45 °C, 0–5% and 6.5–8, respectively. Sequencing and PCR amplification of 16S rRNA gene sequences as a phylogenetic tree revealed that this isolate is closely associated with the genus Acintobacter and represents a distinct sub-line within its members (Fig. 3).
Fig. 2b showed that the best metal ion (25 mM) to maximize the enzyme activity by the newly isolated Acinetobacter in a SSF was magnesium (as MgCl2·6H2O, 4181.25 U g−1), followed by calcium (as CaCl2, 3903.5 U g−1), sodium (as NaCl, 3482.5 U g−1), manganese (as MnCl2·4H2O, 3070.0 U g−1), potassium (as KCl, 3070.0 U g−1), copper (as CuCl2, 1431.25 U g−1), and iron (as FeCl3·6H2O, 566.25 U g−1). The PB design revealed that MCS was the solid substrate better than grape and pomegranate seeds (Table 1). As earlier mentioned, the process parameters of agitation rate and MC were more effective than the particle size according to the PB design.
Optimization study
The predicted model for LA was highly significant (p < 0.0001, Table 3). There was a non-significant lack of fit that further validates the model (p > 0.05). The values of the R2 (0.975) and Radj2 (0.953) also confirmed that the model was highly significant. Adequate precision compares the range of the predicted values at the design points to the average prediction error. This factor measures the signal-to-noise ratio so that a ratio greater than 4 is desirable.8 For the proposed model, this value was 19.74, a very good signal-to-noise ratio. For the LA, effect of MCS/yeast extract (X1), MC (X3) and agitation rate (X4) was significant (p < 0.05) in first-order linear effect, second-order quadratic effect (X12, X22, X32 and X42) and interactive effect (X1X2, X1X3, X1X4, X2X3 and X3X4) (Table 3). The variables with the largest effect were the linear terms of MCS/yeast extract and agitation rate followed by the quadratic effect of agitation rate. The three-dimensional (3D) surface plot was drawn to visualize the significant (p < 0.05) interaction effect of independent variables on the LA (Fig. 4). As considered in Fig. 4, the LA considerably increased as the MC was decreased, but increased with increasing rotation rate. Moreover, an decrease in concentration of olive oil and MCS/yeast extract provided high activity value for the produced lipase. The optimum location led to the highest LA (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480.2 U g−1) for the model was estimated to be achieved by a set level of 4.0 w/w, 30 g L−1, 65.0% and 180.0 rpm for MCS/yeast extract, olive oil concentration, MC and agitation rate, respectively. The suitability of the presented model for predicting the optimum value of LA was delaminated using the recommended optimum conditions. The maximum activity by lipase synthesized experimentally was found to be 20
480.2 U g−1) for the model was estimated to be achieved by a set level of 4.0 w/w, 30 g L−1, 65.0% and 180.0 rpm for MCS/yeast extract, olive oil concentration, MC and agitation rate, respectively. The suitability of the presented model for predicting the optimum value of LA was delaminated using the recommended optimum conditions. The maximum activity by lipase synthesized experimentally was found to be 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 892.7 U g−1, which was clearly very close to the predicted value (p > 0.05).
892.7 U g−1, which was clearly very close to the predicted value (p > 0.05).
| Source | Coefficient | Sum of squares (SS) | dfa | Mean of squares | F-value | Probability > Fb,c,d |
|---|---|---|---|---|---|---|
| a df: degree of freedom.b Significant (*p < 0.05).c Highly significant (**p < 0.01).d ns: not significant. | ||||||
| Model | 3387.00 | 9.807 × 108 | 14 | 7.005 × 107 | 45.24 | <0.0001** |
| X1 (MCS/yeast extract) | −2829.35 | 1.921 × 108 | 1 | 1.921 × 108 | 124.07 | <0.0001** |
| X2 (olive oil concentration) | — | 5.592 × 106 | 1 | 5.592 × 106 | 3.61 | ns |
| X3 (MC) | −2023.19 | 9.824 × 107 | 1 | 9.824 × 107 | 63.44 | <0.0001** |
| X4 (agitation rate) | 2596.90 | 1.619 × 108 | 1 | 1.619 × 108 | 104.52 | <0.0001** |
| X11 | 1709.53 | 8.357 × 107 | 1 | 8.357 × 107 | 53.97 | <0.0001** |
| X22 | 1835.40 | 9.633 × 107 | 1 | 9.633 × 107 | 62.21 | <0.0001** |
| X33 | 2057.53 | 1.211 × 108 | 1 | 1.211 × 108 | 78.17 | <0.0001** |
| X44 | 2273.65 | 1.478 × 108 | 1 | 1.478 × 108 | 95.46 | <0.0001** |
| X12 | 1336.03 | 2.856 × 107 | 1 | 2.856 × 107 | 18.44 | 0.0006** |
| X13 | −1146.09 | 2.102 × 107 | 1 | 2.102 × 107 | 13.57 | 0.0020** |
| X14 | −1510.59 | 3.651 × 107 | 1 | 3.651 × 107 | 23.58 | 0.0002** |
| X23 | −2087.47 | 6.972 × 107 | 1 | 6.972 × 107 | 45.02 | <0.0001** |
| X24 | — | 1.935 × 105 | 1 | 1.935 × 105 | 0.12 | ns |
| X34 | −1244.34 | 2.477 × 107 | 1 | 2.477 × 107 | 16.00 | 0.0010** |
| Residual | — | 2.478 × 107 | 16 | 1.549 × 106 | — | — |
| Lack of fit | — | 1.873 × 107 | 10 | 1.873 × 106 | 1.86 | 0.2312ns |
| Pure error | — | 6.045 × 106 | 6 | 1.007 × 106 | — | — |
| Core total | — | 1.005 × 109 | 30 | — | — | — |
| R2 = 0.975; adjusted-R2 = 0.953; CV = 13.12 adequate precision = 19.74 | ||||||
 | ||
| Fig. 4 3D surface plots showing the significant (p < 0.05) interaction effects on the activity variation of lipase (a–e) produced by Acinetobacter sp. isolated from CSW. | ||
Purification study
The produced lipase by Acintobacter isolated from CSW was purified using ammonium sulphate precipitation (ASP) followed by HIC. In the chromatography, the enzyme was eluted with the buffer which was highly hydrophobic. Purity and activity recovery were increased 26.9-fold and 27.8%, respectively (p < 0.01). Fig. 5a showed the purified lipase is as a single protein band in the SDS-PAGE representing the protein content with high purity level. The molecular mass of the biosynthesized lipase was determined to be about 46 kDa proposing that the enzyme is a monomer. Fig. 5b depicted the FT-IR spectra from 500–2000 cm−1 for the reference lipase and the enzyme produced by the identified isolate before and after the purification process. The closeness of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–N stretching vibrations in these spectra demonstrated that the ASP and HIC can considerably improve purity of the biosynthesized lipase.
O and C–N stretching vibrations in these spectra demonstrated that the ASP and HIC can considerably improve purity of the biosynthesized lipase.
4 Discussion
Rhodamine B-agar plate (RBAP) method was used to determine lipase producing bacteria. It is suggested that the formation of fluorescent halos under the UV rays in RBAP can be due to rhodamine B dimers complexed with mono or diglycerides and fatty acid liberated by the enzyme into the medium.16 High activity of produced lipase by the Acinetobacter isolated from CSW can probably be attributed to the ideal growth conditions and substrate type present in Caspian Sea water.17 Acinetobacter sp. have a high distribution and survival on moist and dry surfaces. A number of lipase producing bacteria especially Acinetobacter sp. inhabit the outer body of many fishes and can feed upon different minerals and protein sources. It has been reported that Acinetobacter sp. are one of the most important lipase-producing microorganisms such as A. calcoaceticus,18 A. radioresistens,19 and A. baylyi.20Minerals play a significant role in production of extracellular lipase. They, as cofactors of several enzymes involved in the biosynthetic pathway of enzymes, can improve valuable metabolites at certain concentrations.21,22 Among the different ions, magnesium and calcium led to a considerable increase in LA by the newly isolated Acinetobacter. Kok et al.23 found that production of extracellular lipase by A. calcoaceticus BD 413 was significantly enhanced when the medium was supplemented with divalent cations of Mg2+, Ca2+, Cu2+ and Co2+. An increase in lipase biosynthesis by A. niger and Pseudomonas pseudoalcaligenes F-111 in the presence of Mg2+ ions was observed.24,25 Hasan et al.3 had reported that Mg2+ ions would generally increase the lipase production by forming complexes with ionized fatty acids, changing their solubility and behavior at interfaces.
The higher effect of MCS than the milled seeds of grape and pomegranate on the LA can be attributed to the high nitrogen content of this seed. It was demonstrated that microorganisms provide high yields of lipase when organic nitrogen sources together with olive oil as carbon source are used at optimal concentrations.23 Moreover, response surface optimization showed that the MCS/yeast extract and agitation rate had more effect than MC and olive oil concentration on the lipase productivity of aerobic microorganism of Acinetobacter in shake flask cultures. These factors at their optimum points can improve substrate's particle structure with the increasing MCS porosity and solubility to transfer oxygen to the isolated Acinetobacter and can increase the LA.26 A higher MC to achieve the highest LA by A. niger (71.0%) was reported than the isolated Acinetobacter (65.0%) in this study.27 However, Mahanta et al.28 found an optimum MC of 50% for the lipase production by P. aeruginosa in SSF using Jatropha curcas seed cake as substrate. The LA reduction by increasing MC can be attributed to the aggregation of substrate particles, poor aeration, and possible anaerobic conditions.6 Similarly, MC lower than optimum level can considerably decrease the solubility and/or degree of swelling of the solid substrate, and thus by providing a higher water tension can produce lipase with a lower activity.26 The LA enhancement by increasing agitation rate can be due to the improved oxygen transfer rate as well as the mixing efficiency of the culture, thus enhancing the cell growth and lipase production. However, higher agitation rate more than 180 rpm by increasing shear force led to a negative impact on cell growth and enzyme activity.29 Alonso et al.30 have also reported 200 rpm, stirring rates as optimum while Freire et al.31 have reported maximum production of lipase at agitation rate of 300 rpm with Pencillium sp. These researchers showed that an increase in agitation rate more than the optimal level (200 or 300 rpm) resulted in decreased enzyme production because of mechanical and/or oxidative stress, excessive foaming, disruption and physiological disturbance of the cells.30,31 Activity reduction of the synthesized lipase at low agitation rates can be attributed to the limited oxygen levels along with the lacking of homogeneous suspension of the fermentation medium and breaking of the clumps of the cells.32 Low increase of the enzyme activity in presence of olive oil inducer in the culture medium could be due to its high content of long, unsaturated fatty acyl chains, such as oleic acid. This fact was earlier demonstrated by other researchers on the lipase production from Amycolatopsis mediterranei and Microbacterium luteolum.17,33 The LA (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480.2 U g−1) obtained at the optimal conditions was considerably more than activity of lipase produced by Yarrowia lipolytica (69.0 U g−1),34 Penicillum sp. (140.7 U g−1),35 Rhizopus oryzae (96.52 U g−1),36 A. radioresistens (54 U L−1),37 Bacillus sp. (168 U mL−1),38 A. niger (33.03 U mL−1),39 Acinetobacter sp. EH28 (57.1 U mL−1),40 Geobacillus thermoleovorans CCR11 (2283 U mL−1),41 A. terreus (1566 U mL−1)42 and P. aeruginosa AAU2 (0.432 U mL−1)43 in SSF cultures.
480.2 U g−1) obtained at the optimal conditions was considerably more than activity of lipase produced by Yarrowia lipolytica (69.0 U g−1),34 Penicillum sp. (140.7 U g−1),35 Rhizopus oryzae (96.52 U g−1),36 A. radioresistens (54 U L−1),37 Bacillus sp. (168 U mL−1),38 A. niger (33.03 U mL−1),39 Acinetobacter sp. EH28 (57.1 U mL−1),40 Geobacillus thermoleovorans CCR11 (2283 U mL−1),41 A. terreus (1566 U mL−1)42 and P. aeruginosa AAU2 (0.432 U mL−1)43 in SSF cultures.
A suitable lipase purity is mostly required for use in the different areas such as food processing, detergent, paper and pulp industry. Therefore, it is necessary to remove some unwanted impurities like proteins because of their antagonistic effects on the desired enzyme's activity.3 Partial purification of the extracted lipase from free fraction of liquid culture by ASP and HIC led to a considerable increase in the purity (26.9-fold) in comparison to other reports. Ahmed et al.40 and Lee et al.44 using the same methods respectively achieved 24.2-fold and 9-fold purification for lipases produced by Acinetobacter sp. EH28 and Acinetobacter ES-1. Uttatree et al.20 reported a successful purification for lipase synthesized by A. baylyi (21.89-fold) to homogeneity by ASP and gel-permeable column chromatography with a relative molecular mass of 30 kDa. The enzyme recovery (27.8%) by the studied Acinetobacter was higher than the recovery of lipase biosynthesized by Geotrichum sp. SYBC WU-3 (20.4%) and Ralstonia sp. CS274 (20.8%).45,46 However, the low recovery may be attributed to strong affinity of the produced lipase with the matrix which may hold some activity even using highly hydrophobic elution condition.47 The molecular mass of lipase produced by newly isolated Acinetobacter (∼46 kDa) was found in the range of other lipases synthesized from the same genus such as A. colcoaceticus BD413 (32 kDa), A. radioresistens CMC-1 (45 kDa), A. calcoaceticus LP009 (23 kDa), and Acinetobacter sp. RAG-1 (43 kDa).4,5,18,20 IR radiation is adsorbed by the polypeptide chain backbone in IR spectroscopy and excited the vibrational modes of basic amide (I and II) bonds to determine protein secondary structure. Main protein bonds because of the peptide group vibration occurred in the spectral region of 1000–1700 cm−1.47 The signals at 1645 cm−1 in the partial purified sample is due to the C–O stretching vibrations of amide I. The bonds at 675, 1111, 1412 and 3390 cm−1 are respectively assigned to C–N stretching vibrations of amide I, partly wide peak of amide I, bending peak of amide I and O–H functional group. Comparison of between infrared absorption spectrum of the purified and crude enzymes revealed that the bands at 1659, 1393, 663, 1077 and 3436 cm−1 in the crude enzyme can be due to the C–O stretching vibrations of amide I, C–N stretching vibrations of amide I, partly wide peak of amide I, bending peak of amide I and O–H group, respectively. In the purified enzyme sample, the band at 1545.5 cm−1 is also assigned to the N–H bending (amide II region) with a contribution of the C–N stretching vibrations.47,48 Moreover, the band at 1234.7 cm−1 in this sample can be ascribed to the N–H bending and, C–C and C–N stretching vibrations that this peak shows random coil conformation of proteins. The amide I band at 1645.63 cm−1 is due to the stretching vibrations of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds in the backbone of the protein; therefore, the frequency of this peak is sensitive to protein secondary structure.48
O bonds in the backbone of the protein; therefore, the frequency of this peak is sensitive to protein secondary structure.48
5 Conclusion
The microbial lipase production by a new strain of Acinetobacter sp. using milled seeds of coriander and olive oil in batch SSF flasks was studied. The obtained results are promising because this strain produces lipase with the highest activity (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480.2 U g−1) in an inexpensive medium which facilitates its recovery and purification. Partial purification led to an increase in the purity (26.9-fold) and activity recovery (27.8%). The biosynthesized lipase from Acinetobacter isolated from CSW will be exposed for its potential esterification and transesterification reaction in organic solvents, synthesis of some industrial esters and biotreatment of lipid-rich industrial wastewaters. However, further studies are needed to purify and characterize the produced lipase to utilize in industrial applications.
480.2 U g−1) in an inexpensive medium which facilitates its recovery and purification. Partial purification led to an increase in the purity (26.9-fold) and activity recovery (27.8%). The biosynthesized lipase from Acinetobacter isolated from CSW will be exposed for its potential esterification and transesterification reaction in organic solvents, synthesis of some industrial esters and biotreatment of lipid-rich industrial wastewaters. However, further studies are needed to purify and characterize the produced lipase to utilize in industrial applications.
Acknowledgements
The authors gratefully acknowledge the financial support from the University of Tehran and Iranian center of excellence for application of modern technologies for producing functional foods and drinks.References
- K. E. Jaeger, B. W. Dijkstra and M. T. Reetz, Annu. Rev. Microbiol., 1999, 53, 315–351 CrossRef CAS PubMed.
- F. Hasan, A. A. Shah and A. Hameed, Enzyme Microb. Technol., 2006, 39, 235–251 CrossRef CAS PubMed.
- F. Hasan, A. A. Shah and A. Hameed, Biotechnol. Adv., 2009, 27, 782–798 CrossRef CAS PubMed.
- S. J. Chen, C. Y. Cheng and T. L. Chen, J. Ferment. Bioeng., 1998, 86, 308–312 CrossRef CAS.
- E. A. Snellman, E. R. Sullivan and R. R. Colwell, Biochem. Eng. J., 2002, 11, 269–274 Search PubMed.
- A. Pandey and C. R. Soccol, J. Sci. Ind. Res., 2000, 59, 12–22 CAS.
- A. Pandey, C. R. Soccol and D. Mitchell, Process Biochem., 2000, 35, 1153–1169 CrossRef CAS.
- S. M. T. Gharibzahedi, S. H. Razavi and S. M. Mousavi, Carbohydr. Polym., 2013, 96, 21–30 CrossRef CAS PubMed.
- S. M. T. Gharibzahedi, S. H. Razavi and S. M. Mousavi, Czech J. Food Sci., 2014, 32, 326–336 CAS.
- I. Y. Sengun, D. S. NIielsen, M. Karapinar and M. Jakobsen, Int. J. Food Microbiol., 2009, 135, 105–111 CrossRef CAS PubMed.
- J. G. Holt, N. R. Krieg, P. H. A. Sneathm, J. T. Staley and S. T. Williams, in Bergey's Manual of Determinative Bacteriology, Williams and Williams, Baltimore, MD, 9th edn, 1994 Search PubMed.
- F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. A. Seidman, J. G. Smith and D. J. Struhl, in Current Protocol in Molecular Biology, John Wiley and Sons, New York, Unit 24, 1997 Search PubMed.
- W. O. B. Silva, S. Mitidieri, A. Schrank and M. H. Vainstein, Process Biochem., 2005, 40, 321–326 CrossRef PubMed.
- O. H. Lowry, N. J. Rosebrough, A. L. Farr and J. Randal, J. Biol. Chem., 1951, 193, 265–275 CAS.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS.
- C. T. Hou and T. M. Johnston, J. Am. Oil Chem. Soc., 1992, 69, 1088–1097 CrossRef CAS.
- B. Joseph, N. Shrivastava and P. W. Ramteke, J. Genet. Eng. Biotechnol., 2012, 10, 137–144 CrossRef CAS PubMed.
- S. Dharmsthiti, J. Pratuangdejkul, G. Theeragoo and S. Luchai, J. Gen. Appl. Microbiol., 1998, 44, 139–145 CrossRef CAS.
- I. L. Liu and S. W. Tsai, Appl. Biochem. Biotechnol., 2003, 104, 129–140 CrossRef CAS.
- S. Uttatree, P. Winayanuwattikun and J. Charoenpanich, Appl. Biochem. Biotechnol., 2010, 162, 1362–1376 CrossRef CAS PubMed.
- S. M. T. Gharibzahedi, S. H. Razavi and S. M. Mousavi, Eng. Life Sci., 2013, 13, 408–417 CrossRef CAS.
- S. M. T. Gharibzahedi, S. H. Razavi and S. M. Mousavi, RSC Adv., 2013, 3, 23495–23502 RSC.
- R. G. Kok, J. J. V. Thor, I. M. N. Roodzant, M. B. W. Brouwer, M. R. Egmond, C. B. Nudel, B. Vosman and K. J. Hellingwer, Mol. Microbiol., 1995, 15, 803–818 CrossRef CAS PubMed.
- D. Pokorny, J. Friedrich and A. Cimerman, Biotechnol. Lett., 1994, 16, 363–366 CrossRef CAS.
- S. F. Lin, C. M. Chiou, C. Yeh and Y. C. Tsai, Appl. Environ. Microbiol., 1996, 62, 1093–1095 CAS.
- M. B. Veerabhadrappa, S. B. Shivakumar and S. Devappa, J. Biosci. Bioeng., 2014, 117, 208–214 CrossRef CAS PubMed.
- N. D. Mahadik, U. S. Puntambekar, K. B. Bastawde, J. M. Khire and D. V. Gokhale, Process Biochem., 2002, 38, 715–721 CrossRef CAS.
- N. Mahanta, A. Gupta and S. K. Khare, Bioresour. Technol., 2008, 99, 1729–1735 CrossRef CAS PubMed.
- C. H. Liu, C. Y. Chen, Y. W. Wang and J. S. Chang, Biochem. Eng. J., 2011, 58–59, 96–102 CrossRef CAS PubMed.
- F. O. M. Alonso, E. B. L. Oliveira, G. M. D. Ortiz and F. V. P. Meirelles, Braz. J. Chem. Eng., 2005, 22, 9–18 CrossRef CAS PubMed.
- D. M. G. Freire, E. M. F. Teles, E. P. S. Bon and G. L. S. Anna, Appl. Biochem. Biotechnol., 1997, 63, 409–421 CrossRef PubMed.
- M. Elibol and D. Ozer, Process Biochem., 2000, 36, 219–223 CrossRef CAS.
- D. S. Dheeman, G. T. M. Henehan and J. M. Frias, Bioresour. Technol., 2011, 102, 3373–3379 CrossRef CAS PubMed.
- A. Dominguez, M. Costas, M. Longo and A. Sanromán, Biotechnol. Lett., 2003, 25, 1225–1229 CrossRef CAS.
- E. Rigo, J. L. Ninow, M. Di Luccio, J. V. Oliveira, A. E. Polloni, D. Remonatto, F. Arbter, R. Vardanega, D. de Oliveira and H. Treichel, LWT–Food Sci. Technol., 2010, 43, 1132–1137 CrossRef CAS PubMed.
- V. K. Garlapati, P. R. Vundavilli and R. Banerjee, Appl. Biochem. Biotechnol., 2010, 162, 1350–1361 CrossRef CAS PubMed.
- Y. C. Lin, J. Y. Wu and T. L. Chen, Biotechnol. Bioeng., 2001, 76, 214–218 CrossRef CAS PubMed.
- S. Ertuğrul, G. Dönmez and S. Takaç, J. Hazard. Mater., 2007, 149, 720–724 CrossRef PubMed.
- F. J. Contesini, V. C. F. da Silva, R. F. Maciel, R. J. de Lima, F. F. C. Barros and P. de Oliveira Carvalho, J. Microbiol., 2009, 47, 563–571 CrossRef CAS PubMed.
- E. H. Ahmed, T. Raghavendra and D. Madamwar, Bioresour. Technol., 2010, 101, 3628–3634 CrossRef CAS PubMed.
- M. G. Sanchez-Otero, I. I. Ruiz-López, D. E. Ávila-Nieto and R. M. Oliart-Ros, New Biotechnol., 2011, 28, 761–766 CrossRef CAS PubMed.
- B. K. Sethi, J. R. Rout, R. Das, P. K. Nanda and S. L. Sahoo, Ann. Microbiol., 2013, 63, 241–252 CrossRef CAS.
- A. Bose and H. Keharia, Biocatal. Agric. Biotechnol., 2013, 2, 255–266 Search PubMed.
- W. K. Lee, A. H. Bea and H. Y. Lee, Enzyme Microb. Technol., 2006, 38, 443–448 CrossRef PubMed.
- Y. Cai, L. Wang, X. Liao, Y. Ding and J. Sun, Process Biochem., 2009, 44, 786–790 CrossRef CAS PubMed.
- H. Y. Yoo, J. R. Simkhada, S. S. Cho, D. H. Park, S. W. Kim, C. N. Seong and J. C. Yoo, Bioresour. Technol., 2011, 102, 6104–6111 CrossRef CAS PubMed.
- M. van de Weert, P. I. Haris, W. E. Hennink and D. J. Crommelin, Anal. Biochem., 2001, 297, 160–169 CrossRef CAS PubMed.
- A. Barth and C. Zscherp, Q. Rev. Biophys., 2002, 35, 369–430 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2015 |