Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
DOI: 10.1039/C5OB90123E
(Correction)
Org. Biomol. Chem., 2015, 13, 8365-8365
Correction: Biofunction-assisted aptasensors based on ligand-dependent 3′ processing of a suppressor tRNA in a wheat germ extract
Atsushi
Ogawa
* and
Junichiro
Tabuchi
Proteo-Science Center, Ehime University, 3 Bunkyo-cho, Matsuyama, Ehime 790-8577, Japan. E-mail: ogawa.atsushi.mf@ehime-u.ac.jp; Fax: +81 89 927 8450; Tel: +81 89 927 8450
Received
9th July 2015
, Accepted 9th July 2015
First published on 15th July 2015
Abstract
Correction for ‘Biofunction-assisted aptasensors based on ligand-dependent 3′ processing of a suppressor tRNA in a wheat germ extract’ by Atsushi Ogawa et al., Org. Biomol. Chem., 2015, 13, 6681–6685.
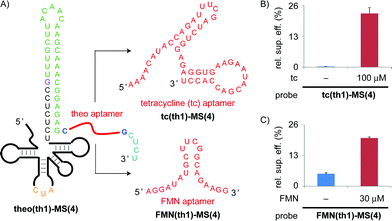
The authors regret that there was an error in the sequence of the FMN aptamer in Fig. 5A. The correct figure is shown below.
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
| This journal is © The Royal Society of Chemistry 2015 |