Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Generation of long, fully modified, and serum-resistant oligonucleotides by rolling circle amplification†
Marcel
Hollenstein
Department of Chemistry and Biochemistry, University of Bern, Freiestrasse 3, CH-3012 Bern, Switzerland. E-mail: hollenstein@dcb.unibe.ch; Fax: (+41) 31-631-3422
First published on 7th August 2015
Abstract
Rolling Circle Amplification (RCA) is an isothermal enzymatic method generating single-stranded DNA products consisting of concatemers containing multiple copies of the reverse complement of the circular template precursor. Little is known on the compatibility of modified nucleoside triphosphates (dN*TPs) with RCA, which would enable the synthesis of long, fully modified ssDNA sequences. Here, dNTPs modified at any position of the scaffold were shown to be compatible with rolling circle amplification, yielding long (>1 kb), and fully modified single-stranded DNA products. This methodology was applied for the generation of long, cytosine-rich synthetic mimics of telomeric DNA. The resulting modified oligonucleotides displayed an improved resistance to fetal bovine serum.
Introduction
The intricate and enzyme-based in vivo DNA replication machinery has been a source of inspiration for the generation of strategies for the in vitro amplification of nucleic acids. In this context, the polymerase chain reaction (PCR) and rolling circle amplification (RCA) are the most used and renowned methods for the amplification of DNA.1–3 RCA is an isothermal polymerase-mediated replication of single-stranded (ss) DNA nanocircle templates that yields long ssDNA products containing numerous repeats complementary to the sequence of the circular precursor.4–8 This amplification tool has found a variety of applications including DNA quantitation,6,9 sensing of biologically relevant analytes,10–14 whole genome analysis,3 and the construction of DNA nanowires.15 Furthermore, small circular DNAs have served as templates for the generation of synthetic mimics of human telomeres.16,17Due to staggering advances in organic synthesis and polymerase engineering, the co-polymerization of modified deoxynucleoside triphosphates (dN*TPs) has advanced as a versatile and convenient method for increasing the chemical repertoire of the functionality deprived nucleic acids.18,19 Indeed, the inclusion of dN*TPs in primer extension reactions has allowed to equip nucleic acids with a variety of functional groups including amino acids,20–24 redox labels,25–27 side-chains capable of organocatalysis,28 transition metals,29,30 boronic acids,31 and even oligonucleotides.32 These modified nucleotides are typically inserted in oligonucleotides by primer extension reactions (PEX) or PCR, yielding dsDNA products which can then be further converted to ssDNAs. Surprisingly, while dN*TPs have been engaged in a multitude of practical applications including in vitro selections of catalytic nucleic acids,33,34 only few examples report on the combined use of dN*TPs and RCA, which allows for the generation of modified ssDNA products. In this context, single dN*TPs (mainly dUTPs) adorned with fluorophores including DY-555 and Cy5 have been used in RCA for fluorescence imaging purposes of the ensuing reaction products,35–39 while a dATP equipped with a biotin residue was recently included in RCA in order to facilitate the labelling of the long ssDNA products with ZnS nanocrystal clusters.40 Finally, Marx et al. took advantage of the combined use of modified dUTPs and RCA to generate long DNAs embellished with short organic polymers.41 All these examples involve the inclusion of single, base-modified dNTPs, mainly mediated by the Φ29 DNA polymerase, which is rather intolerant to modified substrates.38,42 However, some applications would benefit from the inclusion of multiple and differentially modified dNTPs, and thus a general method for the insertion of chemical diversity into RCA products would be an interesting asset.
Herein, I report that deoxynucleoside triphosphates modified at any possible site, i.e. at the level of the nucleobase, the backbone, and the sugar unit (Fig. 1), are fully compatible with the rolling circle amplification method. Application of this strategy allowed for the generation of very long (>1 kb) single-stranded and highly functionalized DNAs. Moreover, dN*TPs could be employed to generate long, cytosine-rich mimics of telomeric DNA. The combination of RCA with dN*TPs represents a potentially useful method for the generation of long fully modified ssDNAs with increased resistance to nucleases, which might be engaged in a variety of potent applications including biosensing,4,43 generation of nanomaterials,15,39 and the development of more potent catalytic nucleic acids.14,44
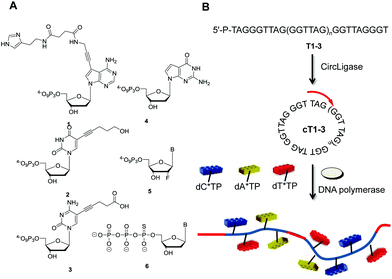 | ||
| Fig. 1 (A) Chemical structure of the modified dN*TPs used to test the compatibility with RCA (B = A, C, T); (B) schematic representation of the generation of long, modified ssDNAs with rolling circle amplification. The templates consisting of G-rich telomeric repeats (for T1–3, n = 4–6)45 are circularized with CircLigase™. In the presence of a suitable primer, a DNA polymerase, and dN*TPs, the nanocircle DNAs serve as virtually infinite templates in RCA. | ||
Results and discussion
Design of the templates and dN*TPs
With the intent of investigating on the compatibility of dN*TPs with RCA, the DNA nanocircle templates cT1–3 developed by Kool et al. were chosen (Fig. 1B).45 These templates consist of 7–9 perfect G-rich telomeric repeats (GGGTTA) and are ideally suited for the generation of long C-rich products. These DNA nanocircles also allowed for the assessment of the compatibility of dN*TPs modified at every possible location (viz. the nucleobase, the sugar, or the backbone) on templates with a more demanding topology. The circular templates cT1–3 are accessible only by application of an orthogonal protecting group strategy at the level of the linear precursors T1–3, since the splint oligonucleotides are prone to hybridize only in the center of these sequences.17,45,46 However, this additional synthetic step can be circumvented by using CircLigase™, an ATP-dependent ligase that mediates the intramolecular ligation of 5′-phosphorylated single-stranded oligonucleotides.47,48 Indeed, when the unprotected 5′-phosphorylated oligonucleotide precursors are incubated at 60 °C in the presence of CircLigase™, only the circular products are formed (ESI, Fig. S1†), which could be isolated in acceptable yields after gel elution (15–40% isolated yields). The integrity of the formed circular products was assessed by exonuclease I digestion (ESI, Fig. S2†).Polymerase screen under RCA conditions with the G-rich templates cT1–3
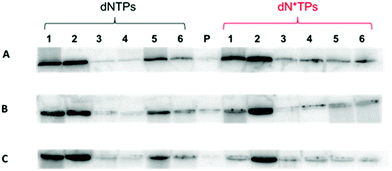
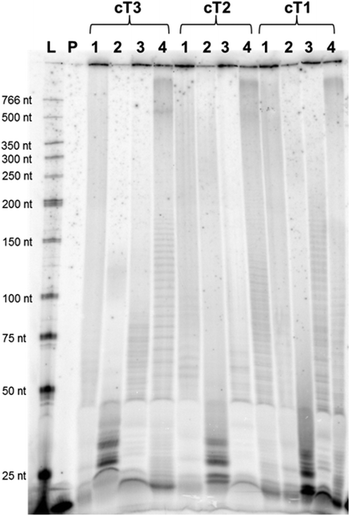
With the circular templates in hand, a panel of DNA polymerases (both A- and B-family) was screened to assess the ability of extending a primer under RCA conditions in the presence of both unmodified and modified dNTPs (Fig. 1B). Concerning the dN*TPs, triphosphates 1–3 bearing amino acid like side-chains anchored at C5 of the pyrimidine and N7 of the purine nucleobases,24 sugar-modified 2′-fluoro-rNTPs 5, and backbone-modified α-thio-dNTPs 6 were chosen as substrates (Fig. 1A). The polymerization reactions with template cT3 and primer P1 followed standard procedures41,45 and analyses of the resulting products are shown in Fig. 2 and the ESI (Fig. S3–5†). Various polymerases including Vent (exo−), 9°Nm and even Pfu were capable of efficiently extending the primer to generate long, single-stranded products with natural dNTPs (lanes 1, 2, and 5, respectively). On the other hand, only the 9°Nm DNA polymerase seemed to be proficient at the polymerization of the dN*TPs under these conditions (lanes 2 in Fig. 2). As observed in a previous report,45 the Φ29 DNA polymerase displayed little propensity at extending the primer P1 under RCA conditions in the presence of either modified or natural dNTPs (lanes 4). Similar results were also observed with the other slightly shorter templates cT1 and cT2, both in the presence of single and multiple dN*TPs in the reaction mixtures (ESI, Fig. S6–12†). Moreover, using 9°Nm as the polymerase leads to the conversion of the majority of the primer P1 into long (>1 kb) products regardless of the nature of the modification (Fig. 3). As estimated by quantitative autoradiography (ESI, Table S1†), the α-thio dNTPs 6 are most efficiently incorporated (∼30% of conversion of the primer to RCA product), followed by the base-modified dNTPs 1–3 (∼20%), and the natural triphosphates (∼10%). Not surprisingly, the 2′-fluoro-rNTPs 5 led to the lowest conversion (∼5%) since they are rather moderate substrates for DNA polymerases with the exception of 2′F-rCTP.49,50In order to exclude the possibility that the observed products stem from another mechanism than rolling circle (e.g. multiround rolling hairpin replication7), primer extension reactions (PEX) were carried out with the linear template T1 and the 9°Nm DNA polymerase (ESI, Fig. S13†). The main products observed correspond to fully extended primer P1 with a retardation in the gel mobility of bands in the case where modified dN*TPs were used.24,41 Multiple bands also arose due to the existence of various primer binding sites on each template. On the other hand, very little (<10%) of long products that were not resolved by 10% PAGE, formed under PEX conditions with the linear templates. Moreover, a Sanger sequencing experiment based on dideoxynucleoside triphosphates (ddNTPs) was carried out on the RCA product stemming from the reaction of the 9°Nm DNA polymerase with template cT1, primer P1, and natural dNTPs (ESI, Fig. S14†). This sequencing experiment clearly revealed the expected pattern of a product resulting from a rolling circle amplification, thus showing that RCA is the predominating mechanism under these conditions, in agreement with earlier findings.41
RCA with different templates and dN*TPs
Next, the generality of the method was assessed by changing the topology of the templates as well as the nature of the functional groups present on the dN*TPs. In this context, the circular template cT4 (ESI†) containing a randomized N20 region,7 similar to templates used in in vitro selections,18 was chosen to maximize chemical and sequence diversity. Therefore, when dN*TPs 1–3 along with 7-deaza-dGTP 4 were used in conjunction with cT4 and the corresponding primer P2, fully modified RCA products could be obtained with the 9°Nm DNA polymerase (ESI, Fig. S15 and 16, and Tables S2 and S3†). Similarly, when dN*TPs 5 and 6 were used either with natural or modified dGTP 4, long, modified RCA products were consistently observed when cT4 served as the template (data not shown). Also, circular template cT5, which contains the reverse complement of the sequence corresponding to the RNA-cleaving DNAzyme 10–23,51 was obtained by a T4 DNA ligase mediated splint ligation (ESI†). cT5 was then used as a template for RCA with one lone dN*TP or a combination of different dN*TPs along with primer P2 (ESI, Fig. S17†). In all cases, the 9°Nm DNA polymerase could extend the primer to generate long, modified ssDNA RCA products with high efficiencies.The insertion of other functional groups is also possible by RCA. Indeed, when dUTP analogs bearing side-chains that could promote organocatalysis, i.e. proline-based residues or strong H-bond donors,52,53 were included in the RCA reaction with cT4, along with α-thio-dATP, α-thio-dCTP, and dGTP 4, the expected fully modified, ssDNA products were obtained (ESI, Fig. S18†).
Serum stability of the RCA products
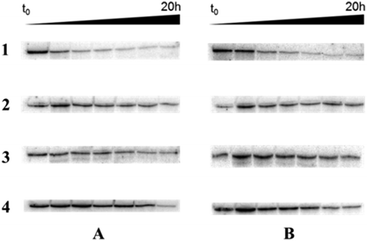
Lastly, the enzymatic stability of the fully modified single-stranded oligonucleotides obtained by RCA in a biological environment was assessed. In this context, the RCA products obtained with template T1 were incubated in 10% heat-deactivated FBS at 37 °C. Fig. 4 shows that little degradation occurred over 10 h with the α-phosphorothioate oligonucleotides (line 3), while the unmodified DNA controls were rapidly degraded under these conditions (line 1). Similarly, the products stemming from the polymerization of the base-modified dN*TPs 1–3 and the 2′-fluoro-rNTPs were rather resilient to degradation (lines 2 and 4, respectively).Conclusions
In summary, I demonstrated that modified deoxynucleoside triphosphates are fully compatible with the rolling circle strategy, regardless of the nature of the modification and the topology of the template. The 9°Nm DNA polymerase, which is more tolerant to modified substrates than the Φ29 DNA polymerase which is typically used for RCA, seems to be the most proficient enzyme for the extension of primers under RCA conditions with dN*TPs. This method was employed to generate fully modified, cytosine-rich mimics of telomeric DNA and all the resulting modified RCA products presented an increased nuclease resistance compared to the unmodified counterparts.The inclusion of chemical diversity into RCA can further broaden the scope of this important methodology and promote the development of new tools for studying biological systems. Moreover, this methodology could have an important impact in synthetic biology, since it guarantees a facile access to fully-modified single-stranded DNA sequences which are difficult to be obtained by any other enzymatic or chemical strategy. Since the inclusion of dN*TPs can enhance the nuclease resistance of the ensuing RCA products new biosensors and biocatalysts can be developed.
Experimental procedures
General procedure for ligation of templates
1 nmol of oligonucleotides T1–T3 were ligated in 40 μL reactions that contained 1× buffer (50 mM MOPS (pH 7.5), 10 mM KCl, 5 mM MgCl2, and 1 mM DTT), 5 mM MnCl2, 100 μM ATP, 1M betaine, and 200 U of CircLigase™. The reactions were incubated at 60 °C for 12 h and then heat deactivated (80 °C, 10 min). The reaction mixtures were precipitated (phenol-chloroform extraction followed by an EtOH wash) and dissolved in 20 μL of H2O. An equal volume of gel loading buffer (formamide (70%), ethylenediaminetetraacetic acid (EDTA; 50 mM), bromophenol (0.1%), xylene cyanol (0.1%)) was added and the ligated oligonucleotides were purified by 20% PAGE (Fig. S1, ESI†). The bands corresponding to product were then eluted (crush and soak method using a buffer consisting of LiClO4 1%, NEt3 1 mM, pH 8), precipitated (EtOH), and desalted (G10 spin columns). 150–400 pmol of circular oligonucleotides were then recovered as determined by UV spectroscopy (A260 measurements).General procedure for exonuclease I treatment54
The cleavage reactions were performed in 20 μL reactions containing 1× reaction buffer (67 mM Glycine-KOH, 6.7 mM MgCl2, 10 mM β-ME, pH 9.5) and 10 U of exonuclease I. The mixtures were incubated at 37 °C for 30 min. Following heat deactivation (80 °C, 10 min), an equal volume of loading buffer was added and the reaction products were analyzed by 15% PAGE (Fig. S2, ESI†).General procedure for rolling circle amplification
The appropriate template (2 pmol per reaction) and 32P-labelled primer (∼1 pmol per reaction) were annealed in the presence of 10× enzyme buffer (provided by the supplier of the DNA polymerase) by heating to 95 °C and then gradually cooling down to room temperature. The adequate DNA polymerase (2 U) and water were then added in turn to the annealed oligonucleotides at 4 °C. The various dNTPs were then added (1 mM final concentration) for a total reaction volume of 20 μL. Following incubation at the optimal temperature for 60 min, the reactions were quenched by adding of 20 μL of stop solution (formamide (70%), ethylenediaminetetraacetic acid (EDTA; 50 mM), bromophenol (0.1%), xylene cyanol (0.1%)). The reaction mixtures were subjected to gel electrophoresis in 5–10% denaturing PAGE containing TBE 1× buffer (pH 8) and 7 M urea. The products were run against a 32P-radiolabelled low molecular weight DNA ladder (units are given in base pairs) that served as a size standard. Visualization was performed by phosphorimaging.Serum stability assays55
The RCA products were generated as described above. The resulting reaction mixtures were heat deactivated (95 °C, 10 min). 4 μL aliquots were then incubated in 10% FBS at 37 °C for given amounts of time. The reaction mixtures were then quenched with 10 μL of stop solution and analyzed by 15% PAGE. The following time-points were taken: 0, 2, 4, 6, 8, 12, and 20 h.Acknowledgements
I would like to gratefully acknowledge Prof. C. Leumann for a critical assessment of this manuscript and for providing lab space and equipment, as well as for his constant support. Dr. Hilda Hollenstein is acknowledged for fruitful discussions. This work was funded by the Swiss National Science Foundation (grant numbers PZ00P2_126430/1 and PZ00P2_144595).Notes and references
- J. Van Ness, L. K. Van Ness and D. J. Galas, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4504–4509 CrossRef CAS PubMed.
- B. Schweitzer and S. Kingsmore, Curr. Opin. Biotechnol., 2001, 12, 21–27 CrossRef CAS.
- R. Johne, H. Müller, A. Rector, M. Van Ranst and H. Stevens, Trends Microbiol., 2009, 17, 205–211 CrossRef CAS PubMed.
- M. M. Ali, F. Li, Z. Zhang, K. Zhang, D. K. Kang, J. A. Ankrum, X. C. Le and W. Zhao, Chem. Soc. Rev., 2014, 43, 3324–3341 RSC.
- W. Zhao, M. M. Ali, M. A. Brook and Y. Li, Angew. Chem., Int. Ed., 2008, 47, 6330–6337 CrossRef CAS PubMed.
- P. M. Lizardi, X. Huang, Z. Zhu, P. Bray-Ward, D. C. Thomas and D. C. Ward, Nat. Genet., 1998, 19, 225–232 CrossRef CAS PubMed.
- A. Fire and S.-Q. Xu, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 4641–4645 CrossRef CAS.
- D. Liu, S. L. Daubendiek, M. A. Zillman, K. Ryan and E. T. Kool, J. Am. Chem. Soc., 1996, 118, 1587–1594 CrossRef CAS PubMed.
- F. Dahl, J. Banér, M. Gullberg, M. Mendel-Hartvig, U. Landegren and M. Nilsson, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 4548–4553 CrossRef CAS PubMed.
- D. A. Di Giusto, W. A. Wlassoff, J. J. Gooding, B. A. Messerle and G. C. King, Nucleic Acids Res., 2005, 33, e64 CrossRef PubMed.
- E. J. Cho, L. Yang, M. Levy and A. D. Ellington, J. Am. Chem. Soc., 2005, 127, 2022–2023 CrossRef CAS PubMed.
- X. Zhu, H. Xu, H. Zheng, G. Yang, Z. Lin, B. Qiu, L. Guo, Y. Chi and G. Chen, Chem. Commun., 2013, 49, 10115–10117 RSC.
- M. M. Ali and Y. Li, Angew. Chem., Int. Ed., 2009, 48, 3512–3515 CrossRef CAS PubMed.
- S. A. McManus and Y. Li, J. Am. Chem. Soc., 2013, 135, 7181–7186 CrossRef CAS PubMed.
- Y. Ma, H. Zheng, C. Wang, Q. Yan, J. Chao, C. Fan and S.-J. Xiao, J. Am. Chem. Soc., 2013, 135, 2959–2962 CrossRef CAS PubMed.
- U. M. Lindström, R. A. Chandrasekaran, L. Orbai, S. A. Helquist, G. P. Miller, E. Oroudjev, H. G. Hansma and E. T. Kool, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 15953–15958 CrossRef PubMed.
- J. S. Hartig and E. T. Kool, Nucleic Acids Res., 2004, 32, e152 CrossRef PubMed.
- M. Hollenstein, Molecules, 2012, 17, 13569–13591 CrossRef CAS PubMed.
- M. Hocek, J. Org. Chem., 2014, 79, 9914–9921 CrossRef CAS PubMed.
- K. Sakthivel and C. F. Barbas, Angew. Chem., Int. Ed., 1998, 37, 2872–2875 CrossRef CAS.
- S. Jäger, G. Rasched, H. Kornreich-Leshem, M. Engeser, O. Thum and M. Famulok, J. Am. Chem. Soc., 2005, 127, 15071–15082 CrossRef PubMed.
- V. Raindlová, R. Pohl and M. Hocek, Chem. – Eur. J., 2012, 18, 4080–4087 CrossRef PubMed.
- Y. Mori, S. Ozasa, M. Kitaoka, S. Noda, T. Tanaka, H. Ichinose and N. Kamiya, Chem. Commun., 2013, 49, 6971–6973 RSC.
- M. Hollenstein, Org. Biomol. Chem., 2013, 11, 5162–5172 CAS.
- M. Hocek and M. Fojta, Chem. Soc. Rev., 2011, 40, 5802–5814 RSC.
- P. Ménová, H. Cahová, M. Plucnara, L. Havran, M. Fojta and M. Hocek, Chem. Commun., 2013, 49, 4652–4654 RSC.
- J. Balintová, J. Špaček, R. Pohl, M. Brázdová, L. Havran, M. Fojta and M. Hocek, Chem. Sci., 2015, 6, 575–587 RSC.
- M. Hollenstein, Chem. – Eur. J., 2012, 18, 13320–13330 CrossRef CAS PubMed.
- L. Kalachova, R. Pohl, L. Bednárová, J. Fanfrlík and M. Hocek, Org. Biomol. Chem., 2013, 11, 78–89 CAS.
- L. Kalachova, R. Pohl and M. Hocek, Org. Biomol. Chem., 2012, 10, 49–55 CAS.
- Y. Cheng, C. Dai, H. Peng, S. Zheng, S. Jin and B. Wang, Chem. – Asian J., 2011, 6, 2747–2752 CrossRef CAS PubMed.
- A. Baccaro, A.-L. Steck and A. Marx, Angew. Chem., Int. Ed., 2012, 51, 254–257 CrossRef CAS PubMed.
- M. Hollenstein, C. J. Hipolito, C. H. Lam and D. M. Perrin, ACS Comb. Sci., 2013, 15, 174–182 CrossRef CAS PubMed.
- A. I. Taylor, V. B. Pinheiro, M. J. Smola, A. S. Morgunov, S. Peak-Chew, C. Cozens, K. M. Weeks, P. Herdewijn and P. Holliger, Nature, 2015, 518, 427–430 CrossRef CAS PubMed.
- I. V. Smolina, D. I. Cherny, R. M. Nietupski, T. Beals, J. H. Smith, D. J. Lane, N. E. Broude and V. V. Demidov, Anal. Biochem., 2005, 347, 152–155 CrossRef CAS PubMed.
- L. Linck and U. Resch-Genger, Eur. J. Med. Chem., 2010, 45, 5561–5566 CrossRef CAS PubMed.
- L. Linck, P. Kapusta and U. Resch-Genger, Photochem. Photobiol., 2012, 88, 867–875 CrossRef CAS PubMed.
- L. Linck, E. Reiss, F. Bier and U. Resch-Genger, Anal. Methods, 2012, 4, 1215–1220 RSC.
- G. Zhu, R. Hu, Z. Zhao, Z. Chen, X. Zhang and W. Tan, J. Am. Chem. Soc., 2013, 135, 16438–16445 CrossRef CAS PubMed.
- J. Yao, K. Flack, L. Ding and W. Zhong, Analyst, 2013, 138, 3121–3125 RSC.
- A. Baccaro and A. Marx, Chem. – Eur. J., 2010, 16, 218–226 CrossRef CAS PubMed.
- J. A. Esteban, M. Salas and L. Blanco, J. Biol. Chem., 1993, 268, 2719–2726 CAS.
- D.-L. Ma, D. S.-H. Chan, B. Y.-W. Man and C.-H. Leung, Chem. – Asian J., 2011, 6, 986–1003 CrossRef CAS PubMed.
- Z. Cheglakov, Y. Weizmann, B. Basnar and I. Willner, Org. Biomol. Chem., 2007, 5, 223–225 CAS.
- J. S. Hartig, S. Fernandez-Lopez and E. T. Kool, ChemBioChem, 2005, 6, 1458–1462 CrossRef CAS PubMed.
- U. M. Lindström and E. T. Kool, Nucleic Acids Res., 2002, 30, e101 CrossRef PubMed.
- T. Blondal, A. Thorisdottir, U. Unnsteinsdottir, S. Hjorleifsdottir, A. Ævarsson, S. Ernstsson, O. H. Fridjonsson, S. Skirnisdottir, J. O. Wheat, A. G. Hermannsdottir, S. T. Sigurdsson, G. O. Hreggvidsson, A. V. Smith and J. K. Kristjansson, Nucleic Acids Res., 2005, 33, 135–142 CrossRef CAS PubMed.
- H. Gu, K. Furukawa, Z. Weinberg, D. F. Berenson and R. R. Breaker, J. Am. Chem. Soc., 2013, 135, 9121–9129 CrossRef CAS PubMed.
- C. G. Peng and M. J. Damha, Can. J. Chem., 2008, 86, 881–891 CrossRef CAS.
- T. Kasuya, S. Hori, H. Hiramatsu and T. Yanagimoto, Bioorg. Med. Chem. Lett., 2014, 24, 2134–2136 CrossRef CAS PubMed.
- S. W. Santoro and G. F. Joyce, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 4262–4266 CrossRef CAS.
- M. C. Holland and R. Gilmour, Angew. Chem., Int. Ed., 2015, 54, 3862–3871 CrossRef CAS PubMed.
- X. Fang and C.-J. Wang, Chem. Commun., 2015, 51, 1185–1197 RSC.
- C. M. Tate, A. N. Nuñez, C. A. Goldstein, I. Gomes, J. M. Robertson, M. F. Kavlick and B. Budowle, Forensic Sci. Int.: Genet., 2012, 6, 185–190 CrossRef CAS PubMed.
- A.-B. Gerber and C. J. Leumann, Chem. – Eur. J., 2013, 19, 6990–7006 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: General procedures, oligonucleotide sequences, additional gel images and tables. See DOI: 10.1039/c5ob01540e |
| This journal is © The Royal Society of Chemistry 2015 |