Velocity valleys enable efficient capture and spatial sorting of nanoparticle-bound cancer cells†
Abstract
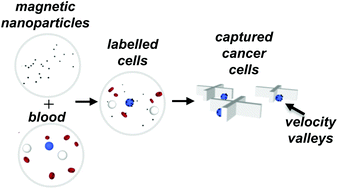
The development of strategies for isolating rare cells from complex matrices like blood is important for a wide variety of applications including the analysis of bloodborne cancer cells, infectious pathogens, and prenatal testing. Due to their high colloidal stability and surface-to-volume ratio, antibody-coated magnetic nanoparticles are excellent labels for cellular surface markers. Unfortunately, capture of nanoparticle-bound cells at practical flow rates is challenging due to the small volume, and thus low magnetic susceptibility, of magnetic nanoparticles. We have developed a means to capture nanoparticle-labeled cells using microstructures which create pockets of locally low linear velocity, termed velocity valleys. Cells that enter a velocity valley slow down momentarily, allowing the magnetic force to overcome the reduced drag force and trap the cells. Here, we describe a model for this mechanism of cell capture and use this model to guide the rational design of a device that efficiently captures rare cells and sorts them according to surface expression in complex matrices with greater than 10 000-fold specificity. By analysing the magnetic and drag forces on a cell, we calculate a threshold linear velocity for capture and relate this to the capture efficiency. We find that the addition of X-shaped microstructures enhances capture efficiency 5-fold compared to circular posts. By tuning the linear velocity, we capture cells with a 100-fold range of surface marker expression with near 100% efficiency and sort these cells into spatially distinct zones. By tuning the flow channel geometry, we reduce non-specific cell adhesion by 5-fold.

 Please wait while we load your content...
Please wait while we load your content...