IRE mRNA riboregulators use metabolic iron (Fe2+) to control mRNA activity and iron chemistry in animals
Elizabeth C.
Theil
ab
aThe Children's Hospital Oakland Research Institute, Oakland, CA 94609, USA. E-mail: etheil@chori.org
bDepartment of Molecular and Structural Biochemistry, North Carolina State University, Raleigh, NC 27695, USA
First published on 11th September 2014
Abstract
A family of noncoding RNAs bind Fe2+ to increase protein synthesis. The structures occur in messenger RNAs encoding animal proteins for iron metabolism. Each mRNA regulatory sequence, ∼30 ribonucleotides long, is called an IRE (Iron Responsive Element), and folds into a bent, A-RNA helix with a terminal loop. Riboregulatory RNAs, like t-RNAs, r-RNAs micro-RNAs, etc. contrast with DNA, since single-stranded RNA can fold into a variety of complex, three-dimensional structures. IRE-RNAs bind two types of proteins: (1) IRPs which are protein repressors, sequence-related to mitochondrial aconitases. (2) eIF-4F, which bind ribosomes and enhances general protein biosynthesis. The competition between IRP and eIF-4F binding to IRE-RNA is controlled by Fe2+-induced changes in the IRE-RNA conformation. Mn2+, which also binds to IRE-RNA in solution, is a convenient experimental proxy for air-sensitive Fe2+ studies of in vitro protein biosynthesis and protein binding. However, only Fe2+ has physiological effects on protein biosynthesis directed by IRE-mRNAs. The structures of the IRE-RNA riboregulators is known indirectly from effects of base substitutions on function, from solution NMR of the free RNA, and of X-ray crystallography of the IRE-RNA–IRP repressor complex. However, the inability to date, to crystallize the free IRE-RNA, and the dissociation of the IRE-RNA–IRP complex when metal binds, have hampered direct identification and characterization of the RNA–metal binding sites. The high conservation of the primary sequence in IRE-mRNA control elements has facilitated their identification and analysis of metal-assisted riboregulator function. Expansion of RNA search analyses beyond primary will likely reveal other, metal-dependent families of mRNA riboregulators.
I. Introduction
Taming iron chemistry in biology is a challenge. A variety of proteins have appeared in animals, plants and bacteria to carry, deliver, scavenge and concentrate iron so it can be safely used in catalysis. In animals, a special class of riboregulators control cellular concentrations of key proteins for cellular iron traffic and use. They are called IRE, Iron Responsive Elements, which are highly conserved nucleotide sequences, folded into bent RNA helix-loop structure. IRE-mRNAs are found in the noncoding regions of animal messenger RNAs (mRNAs).Messenger RNAs share with all, large RNAs, relatively nonspecific Mg2+–RNA interactions because of the long, sugar–polyphosphate backbone. Rapid mRNA turnover, emphasized in the popular Jacob–Monod model of gene regulation,1 is most relevant to mRNA in prokaryotes and the class of mRNAs in eukaryotic that have short half-lives. However, many eukaryotic mRNAs are long-lived (days, weeks), and are “stored” in an inactivate form, until specific molecular signals activate the mRNA for translation and protein biosynthesis.
Historically, metal–RNA interactions have focused heavily on the ionic interactions of magnesium. However, more attention to selective RNA–metal interactions began with the discovery of the metal-dependence of ribozyme; a recent review by the DeRose group is: {Ward, 2014 #376}. Selective metal–RNA interactions are particularly important in mRNAs, because they can amplify the enormous differences in mRNA sequence, size, stability and relative abundance. Moreover, since mRNAs are selectively regulated in general, metal ion–mRNA interactions can contribute to and amplify the regulatory selectivity.
Cell specific regulation may be implemented through metal–mRNA interactions because the mRNA population of particular cell type is a unique mixture of sequences and thus a unique mixture of folded RNA structures. Metal–mRNA complexes will alter the conformations of the mRNA populations of each cell type. By contrast, housekeeping RNAs such as ribosomal and t-RNAs are the same structure and sequence in all living cells, although the quantities of rRNA and t-RNAs can vary among different cells types. In a differentiated cell, each mRNA sequence reflects one of the genes that is differentially transcribed to create the specific protein mixture of a differentiated cell. For example, in cells that are metabolically very active, such as liver hepatocytes, there will be higher concentrations of mRNAs for nuclear-encoded mitochondrial proteins than in fat cells, which have fewer mitochondria. In addition, globin mRNAs are only synthesized in immature red blood cells, while keratin mRNAs are associated with animal epithelial cells and leghemoglobin mRNA is associated with root nodule cells of leguminous plants during nitrogen-fixation. Even in single celled organisms such as bacteria, certain mRNAs are only produced in specific environments. For example Dps protein (mini-ferritin) mRNAs are synthesized when the microbial environment is rich in oxidants such as hydrogen peroxide.3
Since messenger RNAs, like proteins, are single stranded biopolymers, they share many properties. Both RNA and protein biopolymers fold into complex three dimensional structures with loops, bulges and helices, contrasting with the relatively rigid, double-stranded helices of DNA. Folded three dimensional structures in RNA are, as for proteins, sequence-dependent and are stabilized by hydrophobic, ionic and hydrogen-bond interactions. Some RNAs, like some proteins, are chemical catalysts: ribozymes (RNA) and enzymes(protein).2 Rates of synthesis of a particular reflect both the mRNA concentration (DNA transcription and mRNA turnover) and the mRNA activity (translation rate).
The metabolic stability of mRNAs, which varies, is an important regulatory target that complements IRE-riboregulators that control mRNA activity (ribosome binding). For unstable mRNAs, the concentrations of the encoded proteins depend on the degradation rate of the mRNA. Often mRNA turnover depends on specific sequences or structures such as AUREs (AU-rich elements) that are recognized by specific proteins AUF-1, in response to cellular signals and attract ribonucleases4,5 in response to cellular signals. Stable mRNAs, such as most those in the IRE-RNA, by contrast, are inactivated until a biological signal, such as iron, increases the mRNA activity.
MRNAs of the IRE regulatory family contain noncoding mRNA sequences that control the rates of protein synthesis in a set of proteins required for normal iron metabolism and homeostasis; an exception is the transferrin receptor mRNA which combines an AURE (AU-Rich Element) sequence in which five IRE-RNA structures are embedded.6,7 AU-rich RNA elements, as a group, control the stability of many mRNAs by attracting a proteins AUF-1 and nucleases exemplified by globin mRNA in immature red blood cells.8 Thus, the transferrin receptor mRNA, encoding an iron uptake protein, only functions when cellular iron concentrations are low; only then is the IRP binding to the TfR-IRE stable enough to allow significant translation and synthesis of transferrin receptors. But IRE-mRNAs that encode ferritin (iron concentrating and scavenging) or ferroportin (intracellular iron export) only bind IRP when intracellular iron concentrations are high. Only at higher cellular ion concentrations are the stable, IRE-mRNAs able to bind ribosome to allow significant translation and synthesis of ferritin and ferroportin. The IRE mechanism for molecular control of mRNA function still evolving, with the ferritin IRE the oldest, found in primitive organisms such as sponges, while transferrin and ferroportin IRE-RNA structures appeared relatively recently, in vertebrates.7 In a crystal structure of the ferritin IRE-RNA–IRP1 complex, the IRE-RNA folded into a 3D structure that provided multiple contact points with the IRP regulatory proteins (Fig. 1) much like a protein: protein complex.9
IRE-mRNAs are relatively stable with the exception of the TfR mRNA because that IRE structure specifically confers mRNA instability.8 Coincidentally, the location of the TfR, in the 3′UTR, is different than for other IRE-mRNAs, which are in the 5′UTR, but is a relatively common location for signals that regulate mRNA degradation rates. The IRE-RNA regulatory sequences fold into specific 3D structure, which is recognized by a specific regulatory protein called IRP (iron regulatory protein). Most IRE-mRNAs have the IRE-structure in the 5-untranslated (noncoding) region of the mRNA, which is where initiation factors and ribosomes bind. The TfR IRE-RNA structure, which contrasts with most IRE-structures, has a different function. Instead of stabilizing mRNA and increasing the amount of encoded proteins that is synthesized, the TFR IRE structures destabilizes the mRNA and decrease the amount of encoded protein synthesized. The unique features of the TfR mRNA IRE regulatory structure are: (1) TfR IRE sequences are unusually rich in adenine (A) and uracil (U) nucleotides and (2) TfR IRE sequences are located in the 3′UTR, contrasting with most other IRE-mRNAs (the 3′UTR follows the coding region as it its read by the ribosome), contracting with the 5-UTR which precedes the coding regions and facilitates ribosome binding to mRNAs. Both the nucleotide sequence and the position in mRNA of the TfR-IRE RNA are general characteristics of mRNA regulators control mRNA degradation.
The 3′UTR regions of mRNAs that are regulated by degradation are usually rich in A and U nucleotides; such regions are called AUREs (AU-Rich Elements). AUREs attract a protein AUF-1, which, in turn, attracts RNA nucleases that degrade the AURE-targeted mRNA. The cellular conditions that increase initiation factor binding to IRE-RNA in the 5′UTR of IRE-mRNAs and attract proteins such as eIF-4, ribosomes to increase the synthesis of ferritin and ferroportin,10 are the cellular same conditions that attract nucleases to the IRE-RNA structures in TfR (transferring receptor) mRNA and decrease rates of iron entry into the cells. Future studies of the relative binding of IRP and AUF-1 in the presence or absence of iron should be illuminating. Just as Fe2+–RNA binding to IRE riboregulators near the mRNA cap increase initiator factor binding,11 Fe2+ binding to the transferrin receptor IRE riboregulator should increase AUF-1 binding.
IRE-RNA sequences are found, in addition to humans and other vertebrates, in more primitive animals such as sponges, where the IRE regulatory structure is only in ferritin mRNA.7 In higher vertebrates, IRE sequences are also found in mRNAs encoding proteins of with many, different metabolic functions, e.g., for oxidative metabolism (mitochondrial aconitase), oxygen sensitivity (HIF-2α),12 cellular iron export (ferroportin, also IREG-1)13,14 and the synthesis of heme for hemoglobin (erythroid aminolevulinic acid synthase, eALAS),10 as well as for ferritin and other proteins of iron balance. For example, the biosynthesis of mitochondrial aconitase, HIF-2α, ferroportin, and eALAS, like ferritin is regulated by ribosome binding to an mRNA IRE-riboregulator. Apparently the metabolic/reproductive success of organisms that developed mRNA translation sensitivity to iron to complement DNA transcription sensitivity to oxidants, facilitated the spread of IRE riboregulation in evolutionarily more advanced organisms. Whether analogous riboregulator families exist to facilitate cell responses to other, ubiquitous toxic elements, or whether the distinctive roles of iron and oxygen chemistry in biology require the combinations of mRNA regulation with DNA regulation are uncertainties to be resolved in the future.
II. Ferritin DNA, mRNA, protein structure/function – the feedback loop, and other IRE-RNAs
Ferritin DNA and mRNA sequences in plants and animals differ much more in organization than do the encoded protein sequences and of the folded, protein nanocages.15 Ferritin in both plants and animals is encoded in nuclear DNA and synthesized in the cytoplasm. But the functional location of ferritin protein in plant cells is in the subcellular compartment called plastids (amyloplasts, chromoplasts, chloroplasts, etc., depending on the type of plant cell). By contrast, the functional location of ferritin in animals and microbes is the cytoplasm. As a result, plant ferritin genes need extra information so the ferritin protein can be transported into the plastids found in root, flowers, leaves, etc.; animal ferritins do not need such signals because the functional location of animal ferritin remains in the cytoplasm, the site of protein synthesis. Usually the plant ferritin subunits have an extended N-terminal peptide which informs the cells on the plastid location.16 Plant ferritin genes have more introns than animal ferritin genes,15 coincide both with the more complex intracellular distribution and multiple roles for ferritin in plant development, resistance to oxidative stress, concentrating iron and phosphate richness of ferritin minerals.17Serum ferritin in animals, in contrast with tissue ferritins, is secreted by the cells, that synthesize it, into the serum; it usually is iron-free except during iron-overload. As for many serum proteins, the signal for secretion of serum ferritin is postsynthetic glycosylation of the protein. The cell type most likely to synthesize serum ferritin is from the reticuloendothelium. However, in disease states when tissue damage occurs, ferritin from other tissues can also appear in the serum, by nonsecretory mechanisms. No gene encoding serum ferritin has been identified to date. Part of the difficulty is the presence of many ferritin “pseudogenes” in animal DNA. Ferritin pseudogenes may relate to the stability of ferritin mRNA, which provides opportunities for cDNA copies of ferritin mRNA to find their way into the genomic DNA. The small amounts of ferritin in serum make direct study of the glycosylated ferritin difficult and so, serum ferritin remains poorly understood.18 Nevertheless, serum ferritin concentrations are a major marker in clinical medicine, which is widely used to detect iron deficiency, inflammation, and some cancers.19
The IRE–IRP regulatory system is an example of selective translation in which of a subset of cell mRNAs is repressed by the same protein and activated by a common metabolic signal. Examples of eukaryotic mRNA families with noncoding riboregulators are relatively rare compared to other forms of genetic regulation. Because IRE riboregulators are present in multiple mRNAs encoding proteins of iron homeostasis, the synthesis of the group of proteins can be coordinately regulated by controlling mRNA translation rates. The other currently known example of coordinated control through a family of noncoding riboregulatory structures is the SECIS family of mRNAs, which regulate the biosynthesis rates of proteins containing selenocysteine {Kurokawa, 2013 #377}. It is likely that other such families await discovery.
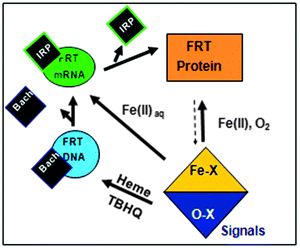
Regulation of mRNA function involves stabilizing mRNA in the cytoplasm and mRNA recognition by specific, cytoplasmic regulatory proteins and metabolic sensors. Such a system consumes cell resources. Possible explanations for the dual regulation of ferritin using both genes (DNA) and ferritin mRNA, on coordination with other iron metabolic genes, include the dual roles of iron in cellular health (iron cofactors) and cell damage (generating reactive oxygen species, ROS). Ferritin DNA is regulated as part of a gene family related to oxidative damage contrasting with the mRNA regulation for iron utilization. The ferritin DNA transcriptional regulator is called ARE (Antioxidant Response Element) and coordinates ferritin mRNA synthesis with a variety of antioxidant proteins.20 Examples are thioredoxin reductase and NADPH-quinone reductase, which return cytoplasmic proteins to their normal redox states after oxidation. ARE regulation of ferritin contributes to recovery from oxidative damage in animal cells by concentrating the iron released from oxidant-damaged iron cofactors such as heme or iron sulfur clusters. The iron concentrated in ferritin is recycled for the biosynthesis of new iron–protein cofactors in the cytoplasm during cell recovery from oxidant damage. Ferritin DNA transcription, and that of other ARE-regulated genes, is repressed by the heme-binding protein, Bach 1.21 Thus, the combination of DNA and mRNA regulation allows ferritin biosynthesis to respond both to the cellular damage iron (oxidative stress) and the activity iron confers on many metabolic proteins.
The relative paucity of mRNA regulation in plants and bacteria could relate to the shorter life expectancies of individual cells in plants and bacteria. Trees, for example, may live for centuries, but individual cells are active for a relatively short time before becoming “woody”. In such cells, transcriptional regulation may be sufficient to maintain iron balance and redox protection. Perhaps frequent cell division combined with random distribution of toxic components during cell division coupled with the extra complexity of coordinating regulation among the nuclear, mitochondrial and plastid genomes, may have overridden some advantages of mRNA regulation. As a result, a functional, plant IRE-RNA may have been lost during evolution. Note that insertion of an animal IRE into a plant ferritin mRNA actually inhibited protein biosynthesis slightly (Y. Kimata, D. R. Dix, M. Ragland, and E. C. Theil, unpublished observations). Support for such a notion is the presence of IRE-cDNA-hybridizing sequences in soybean DNA (M. Ragland, PhD thesis, 1993, North Carolina State University) and the negative effects of soybean mRNA downstream sequences on the function on an inserted, animal, IRE.22,23 Plant genes encoding mRNA with IRE-elements, simply may have been lost during evolution of contemporary plants.
Regulation of ferritin biosynthesis in animals is part of a feedback loop. One component of the feedback loop, ferritin gene DNA transcription (Fig. 1), is sensitive to cellular oxygen signaling and is mediated by the ARE-DNA promoters and the ARE-DNA binding, heme-regulated, protein repressor, Bach 1.20,24 The second component of the feedback loop, ferritin mRNA translation, is sensitive to cellular iron signaling. The third component of the feedback loop is ferritin protein, the gene product, which consumes iron and oxygen to synthesize the protein caged iron-oxy mineral, thereby decreasing “free” iron concentrations and shutting down the feedback loop. Lower oxygen/oxidant concentrations lower ferritin DNA transcription, which lowers ferritin mRNA translation. The iron-induced changes in the activity of ferritin DNA and ferritin mRNA both lower ferritin protein biosynthesis rates. Biological feedback loops are fairly common in biology,25 but the case of ferritin, where the gene product consumes the signals for both DNA and mRNA activation appears to be unique, at least early in the 21st century.
III. Eukaryotic riboregulatory families
There are only two known mRNAs families with noncoding riboregulators, at this time. They are the IRE-RNA family (encoding proteins that balance iron metabolism) and the SECIS RNA family (encoding selenoproteins such as thioredoxin reductase and NADPH quinine reductase).26 No IRE sequences have been found in plant mRNA. No IRE-mRNAs have been found in higher plants. No SECIS RNA had been identified was in higher plants until the recent identification of a SECIS element in the mitochondrial RNA of cranberries.27 SECIS mRNA is also found in the cytoplasm of the single celled, photosynthetic, aquatic organism Chlamydomonas.26,28 A discussion of the distribution of primitive gene distribution in higher plants and Chlamydomonas is found in ref. 29. At this point, the contrast between the evolution of riboregulators in animals, such as IRE mRNAs and SECIS mRNAs,7,26 and the small number of riboregulators in plants, remains unexplained.The sequence similarity among animal ferritin IRE-mRNAs were sufficient for identification, contrasting with some other mRNAs where the same higher order structure is obtained with different sequences; as additional “iron genes” were cloned, more IRE-mRNAs were identified. IRE-RNA, noncoding RNA sequences generally lie near the mRNA ribosome binding site in the 5′UTR;30 the conserved sequence was named IRE, the Iron Responsive Element because of the effect on increasing iron concentrations on the synthesis of proteins encoded in IRE-containing mRNAs. Subsequent studies confirmed the IRE-RNA sequence was required for IRP binding, that the IRE riboregulator occurred in several other mRNAs encoding iron metabolic proteins, and that the IRE riboregulator was required for iron dependent increases in the synthesis of ferritin protein;31 reviewed in ref. 32.
Only two noncoding, mRNA, riboregulatory families are identified to date IRE and SECIS (selenocysteine insertion sequence), there are several possible explanations for their rarity. First, noncoding riboregulators that conserve primary or tertiary structure, but not primary structure (base sequence) are difficult to find with current search methods, since they depend heavily on conservation of linear sequences. Second, mRNA riboregulator families, such as the IRE and SECIS RNAs, appear to be relatively recent in evolution and are still spreading among metabolically related mRNAs in animals. For example, IRE-mRNA appeared first in ferritin, and in invertebrate sponges and later, with a weaker effect (weaker repressor binding) in the mitochondrial (MT) aconitase mRNA of sea squirts, an invertebrate chordate; MT-aconitase is encoded in a nuclear gene, synthesized in the cytoplasm, as is ferritin, and then transported to the mitochondrion, contrasting with ferritin. The transferrin receptor set of IRE-elements and most of the other IRE elements did not appear until later in evolution with the appearance of vertebrates.7 Third, many models of gene regulation with rapid mRNA turnover and predominantly DNA regulation were developed earlier than mRNA regulatory models. For DNA, the ideas began in the middle of the last century1 using single celled organisms, where DNA and mRNA are both readily accessible to cytoplasmic changes. In the nucleated cells of more advanced organisms, DNA is protected from by the nuclear membrane and histone protein, but early model “patched” the prokaryotic biosynthesis onto specialized nucleated cells. Moreover, in multicellular organisms that have highly specialized cells with distinct metabolic needs, gene expression demands more complex coordination and regulation. Finally, recognition of the combination of three-dimensional RNA folding of mRNA before it unfolds/is threaded in the ribosome, and selective binding of regulatory proteins (repressor and activators) is relatively recent.33 The appealing simplicity of the earlier gene regulatory models, and the relative paucity of information about translational mRNA regulation, have both contributed to the slow development of knowledge about mRNA riboregulators in plants and bacteria, as well as in animals. A recent review on plant gene regulation states, “We delineate the need for additional genome-wide studies of RNA secondary structure and RNA–protein interactions in plants”,34 which indicates current awareness of the problem.
Ferritin protein biosynthesis in animals is part of a feedback loop (Fig. 1). One component of the feedback loop, ferritin gene DNA transcription, is sensitive to cellular oxygen signaling. The signals are mediated by the ARE-DNA promoters and the ARE-DNA binding, heme-regulated protein repressor, Bach 1;20,24 signals alter ARE activity differ from the IRE-RNA riboregulators. The signals include H2O2, and other molecules that cause oxidative damage or stress. The second component of the ferritin feedback regulatory loop, is ferritin mRNA translation. IRE-mRNA is activated by direct Fe2+ binding to the IRE-RNA activator.11,35 The third component of the iron feedback loop is ferritin protein, the DNA and mRNA product, which consumes iron and oxygen to make the ferritin protein-caged iron-oxy mineral. As a result, ferritin protein activity consumes the two genetic signals controlling DNA and mRNA activity, which shuts down the feedback loop. Lower oxygen/oxidant concentrations lower ferritin DNA transcription and lower iron concentrations lower ferritin mRNA translation. The combined effects decrease ferritin protein biosynthesis. Biological feedback loops are fairly common,25 but the case of ferritin, where the gene product consumes the signals for both DNA and mRNA activation, the feedback loop appears to be unique, at least now, early in the 21st century.
IV. IRE-RNA structure/function
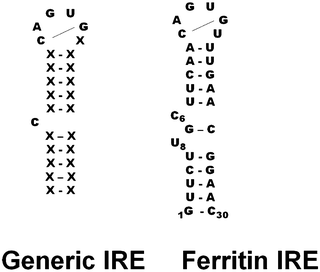
Each member of the IRE riboregulator family has two types of conserved information: information shared by all IRE-mRNAs and information that is IRE-mRNA specific. All IRE-mRNAs have a short (9–10 base pairs), double-strand helix, with an unpaired base C, near the middle of the helix that creates a bulge (Fig. 2 and 3). In analogy to protein helices, the “bulges” or unpaired bases in RNA helices are like amino acids like proline, which interrupt protein helices. The IRE-sequence differences between different mRNAs are relatively small in the sense that all IRE-RNAs form the RNA A-helix with the same bulge C and terminal loop sequence, CAGUGX (shown in bold font in a human ferritin IRE-RNA sequence). In all IRE-RNA, a conserved, C–G base pair across the IRE terminal loop, creates a triloop, AGU, and a bulge (Fig. 1).9,36,37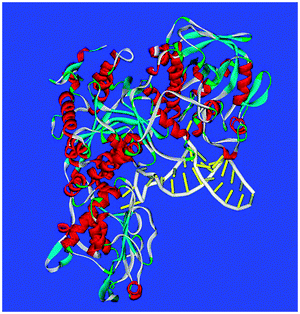 | ||
| Fig. 2 The ferritin IRE-RNA IRP protein complex. PDB file: 2IPY. IRP helices-red; RNA double helix: polyribophosphate backbone-white; paired bases: yellow. PDB file: 2IPY. Note the multiple, intimate RNA–protein interactions along one RNA surface with another RNA surface free for other interactions, such as binding metabolic iron. | ||
Quantitative differences in the cellular concentrations of each IRE-mRNA coincide with quantitative differences in the encoded protein concentrations, under each environmental condition. However, different IRE-RNA–IRP repressor binding stabilities35,38 also contribute to differences in the cellular concentrations of proteins encoded in IRE-mRNAs. To achieve the existing array of IRP binding constants, the RNA sequence conservation of IRE-mRNAs encoding different proteins is lower (80–85%) than the phylogenic conservation of a single IRE-RNA (>90%). The human mt-aconitase and ferritin H IRE mRNAs vary much more in sequence than the ferritin IRE-RNA sequences vary among vertebrates, such as and human, mouse, chicken and frog.7
The regulatory protein that recognizes all IRE-RNA structures is named iron regulatory protein (IRP)35,39 (Fig. 2). Small, conserved differences in IRE-sequence and structure create a family of protein–RNA complexes; each member of IRE-RNA family has a slightly different binding stability of the protein–RNA complex. The IRE-RNA–IRP stability differences have physiological consequences. For example, the Kd (nM) for IRP/ferritin H IRE RNA binding in a solution with 5 μM Fe2+ is: 78.9 ± 4.5, while the Kd for IRP/mt-aconitase under the same conditions is 259 ± 17.35 The RNA targets were the 30 nucleotide IRE-RNA sequences,35 which are embedded in mRNAs hundreds of nucleotides long. Slightly different, absolute Kd values are obtained using shorter IRE-RNA fragments and mobility in electrophoretic gels,39 but the relative differences in IRP binding between different IRE-RNA structures are the same by both techniques.
The result of quantitatively different IRP binding affinities for each different IRE-mRNA,35,40 is that at any one time, the fraction of the IRE-mRNA inactivated by IRP binding will be different for the each IRE-mRNA. The ferritin IRE-RNA, for example forms a much more stable complex with IRP than aconitase – mRNA. As a result ferritin mRNA translation is more resistant to small changes in intracellular iron concentration than mt-aconitase mRNA, which forms a less stable mRNA–IRP complex. Physiologically, since there is a constant cellular need for aconitase in bioenergetics, contrasting with the periodic need for the iron concentrating activities of ferritin, the structural differences in the ferritin mRNA IRE and the mt-aconitase mRNA IRE relate to functional difference in cell metabolism of each protein encoded in an IRE-mRNA.
Structural specificity in the helix sequence of the ferritin IRE-RNA, the oldest IRE-RNA currently known7 is an extra helix bulge below the G–C base pair that closes the generic IRE-RNA C-bulge (Fig. 3).
Differences in mRNA stabilities35,38 also contribute to differences in the cellular concentrations of proteins encoded in IRE-mRNAs. To achieve the existing array of IRP binding constants, the RNA sequence conservation of IRE-mRNAs encoding different proteins is lower (80–85%) than the phylogenic conservation of a single IRE-RNA (90%). The human mt-aconitase and ferritin H IRE mRNAs vary much more in sequence than the ferritin IRE-RNA sequences vary among vertebrates, such as and human, mouse, chicken and frog.7
The result of RNA bulges and base pair differences in RNA helix base pairs among members of the IRE-RNA family (Table 1) is quantitatively different binding of other cellular macromolecules such as IRP repressor or translation initiation factors, or even with the Fe2+ signal itself. The small differences in IRE-RNA structure among different IRE-mRNAs explain why, in vivo, the same amount of iron, in the same tissue, such as liver, increased ferritin protein biosynthesis more than mt-aconitase biosynthesis.41 When cellular iron concentrations are low, a larger fraction of ferritin mRNA molecules are bound to IRP than are mt-aconitase mRNA molecules. The ferritin IRE-RNA–protein dissociation rate is smaller than for mt-aconitase mRNA and the IRE-RNA–protein binding rate is larger for ferritin mRNA than for mt-aconitase IRE-mRNA,10,39,40,42 reflecting metabolic variations in the use of the two encoded proteins. When IRP–IRE-mRNA binding is weakened by increased concentrations of iron, the number of ferritin mRNA molecules that become available for initiation factor and ribosome binding is disproportionately larger than for mt-aconitase mRNAs. The sequence conservation among the IRE-RNA in humans is ∼80% and explains quantitatively difference in iron responses for different proteins encoded in IRE-RNA. In contrast for a single mRNA such as ferritin mRNA, the IRE-mRNA the phylogenetic conservation is >90% mRNAs.7,32
IRE sequence conservation of a particular IRE-mRNA, such as ferritin mRNA among different animals7 but it is much less contrast with the variations among IRE-RNA sequences in different mRNAs of the same animal (Table 1). In humans, for example, the sequence conservation between FPN and FTH (Table 1), is only 10 out 30 nucleotides, 33%, and for mt-aconitase (mtAco) and ferritin H (FTH), only 13/29 bases or 45% are the same. Also clear from Table 1 is that the length of the IRE sequence in each mRNA differs. In general, the longer IRE-RNA sequences have more base pairs in the stem below the bulged C. While all the IRE-RNAs in the 5′UTR inhibit ribosome binding (translation) when iron low and increase translation when iron is high, the IRE-RNAs sequences with a higher affinity for regulatory proteins have a larger quantitative response to iron.10,43
V. Metal IRE–RNA interactions
Metal ions bind to RNA with two different mechanisms. The first mechanism is ionic and reflects the anionic properties of the RNA sugar polyphosphates backbone. Mg2+ is the preferred binding cation. Until relatively recently, this type of ionic binding was the only RNA–metal interaction considered. However, with the increasing attention on small metal dependent RNA catalysts, the peptide bond formation by 50S ribosomal RNA–Mg2+ complexes44 and the growing numbers of RNA crystal structures with metal ions in specific sites, knowledge of metal binding to specific, three dimensional sites in RNA is growing rapidly.33,45 NMR studies in confirmed the predicted IRE-RNA secondary structure, but no metal ions other than Na+, were not present in the solutions {McCallum, 2003 #168}, except a relatively early, low resolution NMR study where cobalt hexamine was observed bound near the internal bulge and the hairpin triloop {Gdaniec, 1999 #167}. Metal–RNA and metal DNA interactions involving Ni2+ and Pd2+ {Ward, 2014 #376} are examples of the vast potential of structural information that could in the future be obtained about the IRE-RNA family. The results of high resolution NMR studies that directly probe the conformational effects of the Fe2+ signaling in RNAs of the IRE-RNA family are experiments that remain for the future.Two types of observations suggest direct Fe2+–IRE RNA binding as the biological mechanisms of iron-dependent regulation of IRE-RNAs. First Fe2+ binds to RNA and changes interactions with the protein repressor, IRP, and RNA conformation.46 Second, Fe2+ activates ferritin mRNA in cell-free protein biosynthesis studies.11,35 Direct binding of Fe2+ binds to IRE-RNA (anaerobic to prevent reactions of Fe2+ with O2) was measured in solution as changes in the fluorescence of IRE-RNA–ethidium bromide complexes;35 the conformational changes in IRE-RNA are independent of the conformation changes in IRP, determined using the fluorescence of the 12 tryptophan residues in IRP as a reporter. In addition, when IRE-RNA was tagged with the fluorescent reporter 2-aminopurine, substituted for A at position 15 (Fig. 1) an IRE-RNA loop/IRP contact site,9 the addition of Fe2+ to IRE-RNA greatly decreased the fluorescence of 2-aminopurine.11
A study of Fe2+ interactions with IRE-RNA in solution, probed with hydroxyl radical that was generated in situ from the reaction of Fe2+ with H2O2, showed unequal RNA cleavage,47 contrasting with the lack of specificity of hydroxyl radical cleavage of other RNAs under the same conditions. The chemistry of RNA cleavage by hydroxyl radicals, generated by Fe2+ and hydrogen peroxide reactions, predict that cleavage will occur at every accessible nucleotide.48 If, however, Fe2+ bound to selective sites on the RNA, than the local concentrations of hydroxyl radical near the bound Fe2+ ions would be much higher than elsewhere giving rise to the uneven intensities of RNA cleavage observed with IRE-RNA.47 Based on such information, the Fe2+ binding site in IRE RNA is near the stem loop bulge formed by U6G7C8 and C25 (Fig. 2), which is paired to G7.49 This is the same region where a number of metal complexes bind, such as 1,10-Cu2+-phenanthroline and [Ru(tpy)(bpy)O]2+.50
Crystallographic analyses, fruitful, as they have been for structural determinations of other RNA–metal complexes,45 cannot provide information about Fe2+ binding to IRE-RNA. First, no IRE-RNA has been crystallized, in part because of inherent conformational flexibility and relatively small size (30 nucleotides). Second, the use of IRP binding protein complexation with IRE-RNA to facilitate crystallization, another approach to crystallizing other RNA–metal complexes,45 cannot be used because metal ions weaken the IRE-RNA–IRP complex.10,35 In fact, the only condition under which an IRE-RNA–IRP complex has been crystallized is without metal ions.9 For these reasons, other physical methods, such as NMR with proxy metal ions for Fe2+ are likely the most effective way to determine the binding site of the metabolic Fe2+ signal in IRE-RNA with current methods.
VI. Fe2+ and cytoplasmic iron effects on IRE-RNA binding to IRP translation repressor protein
Fe2+ effects on protein biosynthesis in vitro mimic the effects of increasing cellular iron concentrations in vivo or in cultured mammalian cell models. Protein biosynthesis is even more complex that DNA biosynthesis or mRNA biosynthesis, since the nucleic acid sequence is not copied.51 Rather, sets of three nucleotides are “translated”/amino acid residue; the three nucleotide code for each amino acid is universal for plants, animals or microbes. The coding triplets in mRNA are bracketed by noncoding, regulatory RNA sequences at the beginning of the mRNA (5′-untranslated region, or 5′UTR) and at the end of or the 3′UTR.The fact that each mRNA is NOT a linear sequence of nucleotides, but in fact, each mRNA is a specifically folded, macromolecular structure, increases the complexity of cellular protein biosynthesis (mRNA translation); the array of all the different proteins in a cell is encoded in a set of different mRNAs varying in nucleotide sequence and three dimensional structures; the single stranded nature if RNA, contrasted with DNA increases the larger number of folding possibilities. However, in contrast to ribosomal RNAs and t-RNAs, and excluding RNA viruses with known structures that are also mRNAs, the 3D structure of mRNAs are unknown.
The structure of Mg2+ complexed to two different mRNA regulatory elements is known: (1) the Mg2+–Internal Ribosome mRNA Entry Site (IRES) complex49 and (2) the Mg2+–catalytic RNA (ribozyme) complex, exemplified by Mg2+ binding to a G–U wobble pair and an GNRA tetraloop.2 For IRE-RNAs, based on changes in RNA-bound, ethidium bromide fluorescence upon Fe2+ binding, the IRE-RNA conformation in the active form (when Fe2+ is bound) is different from free IRE-RNA,11 but more detail awaits future investigations.
Mg2+ has multiple functions at all stages of protein biosynthesis (mRNA translation). By contrast, specific roles of other metal ions in mRNA translation are only beginning to be discovered. An example is Mn2+, which serves an oxygen-resistant proxy for Fe2+. The IRE-mRNA activating effects of Mn2+ are similar to Fe2+in vitro, but Mn2+ has no effect on ferritin synthesis in cell free extracts, while Fe2+ increase ferritin protein synthesis in cell-free extracts.11 However, there is no evidence that Mn2+ regulates ferritin synthesis in vivo. Thus, only Fe2+, and not Mn2+ regulate IRE-mRNAs in vivo.
The known protein types that control IRE-mRNA function are IRP, an IRE-specific regulatory repressor protein and eIF-4F a translation “factor” protein that binds to all mRNAs. There are two IRPs, IRP1 and IRP2. The relative amounts of IRP1 and IRP2 vary among different specialized cell types. IRP RNA repressor proteins are structurally related to mt-aconitase, an iron–sulfur protein that is part of the Krebs cycle (also called the tricarboxylic acid or citric acid cycle) in mitochondria. In fact, IRP1 can bind an iron–sulfur cluster and acquires aconitase activity, a contrast with IRP2. IRP1 will also bind a wider variety of wild type and mutant IRE-RNAs, whereas binding of IRP2 is more selective. Each IRP has phosphorylation sites that control IRP protein turnover.
EIF-4F, Eukaryotic Initiation Factor – 4F is a one of a group of generic proteins (factors) required to begin (Initiate) mRNA translation in organisms with nucleated cells; eIF-4F, is a very large, multisubunit protein that binds both ribosomes and mRNAs; a number of the studies of IRE-RNA/eIF4F and IRP binding were a collaboration with Prof. Dixie J. Goss.52 Fe2+ or Mn2+ weaken IRP–IRE-RNA binding in solution. By contrast metal ions increase the stability of IRE-RNA/eIf-4F binding.11 Thus, IRP and eIF-4F compete for IRE-RNA binding with metal ions driving the binding competition away from IRP and toward eIF4F. The effects are metal ion selective, with Fe2+ and Mn2+ having larger effects than other divalent metal cations such as Mg2+.35 The IRE-RNA–IRP Kd in the absence of divalent cations is 14 nM, at concentrations of 5 μM Fe2+ or Mn2+, the Kd increases to 50–70 nM.35
Mg2+ is present at ∼0.5 mM, physiologically. However, concentration of 2.0 mM Mg2+ are required to change the stability of IRE-RNA–IRP complexes contrasting with 0.05 mM effects for Fe2+.35 Thus, the physiological concentrations of Mg2+, ∼0.5 M, will have few, if any, effects on IRE–IRP interactions, while small changes in the concentrations of Fe2+ can have very large effect on IRE-RNA–IRP dissociation and on ribosome binding to IRE-mRNAs.
Maximum stimulation of ferritin synthesis in vitro (activation of IRE-mRNA) occurred with 0.05 mM Fe2+ or Mn2+,11 contrasting with the 2.0 M Mg2+ required.35 The sensitivity of IRE-mRNA – dependent protein synthesis to Fe2+, or the Mn2+ surrogate, at concentrations far below those of other metal ions present (Mg2+ – 0.5 mM and K+ – 79 mM) emphasizes the metal selectivity of the Fe2+–IRE-RNA interaction (Fig. 3).11 Experiments with IRE-RNA containing the fluorescent reporter 2-aminopurine show that Fe2+ binding to IRE-RNA changes the RNA conformation.11
Fe2+ is much more effective than Mn2+ in controlling ferritin mRNA activity and protein synthesis rates in vivo, which contrasts with the similarity of effects of Mn2+ and Fe2+ on IRE-mRNA–IRP interaction in vitro. Specific Fe2+ transporters that selectively deliver Fe2+ to IRE-RNAs, or differences in the effective concentrations of Mn2+ and Fe2+ in the local vicinity of mRNA–ribosome complexes are likely explanations for the weaker effects of Mn2+ on the synthesis of IRE-mRNA encoded proteins in vivo. Fe2+ binding to IRE-mRNA has opposite effects on two IRE-RNA–protein interactions. When Fe2+ binds to IRE-RNA, IRP repressor dissociates from IRE-RNA and the mRNA is translated into protein such as ferritin, iron uptake protein DMT1 and the iron export protein ferroportin. As IRP dissociates, eIF4F associates with IRE-RNA, presumably because eI4F sites are exposed/created on IRE-RNAs after IRP dissociates. Once eIF4F binds to an IRE-RNA, eIf4F recruits ribosomes to the mRNA, which explains the old observations that excess iron in cells and animals causes IRE-mRNAs to move from cell supernatant fractions (“free” mRNA) to polyribosomes (“bound”/translated mRNA)53,54 “inducing” ferritin synthesis as much as 100-fold.41 The mechanism depends on direct interactions between ferrous ions with IRE-RNAs and correlated changes in IRE-RNA conformation, which make IRP binding less likely.11
Increasing cellular concentrations of iron not only alters IRE-RNA conformation to change IRP and eIf4F binding, but also can the IRP protein itself. For example, when IRP1 binds an FeS cluster and becomes cytosolic aconitase, the ability to bind IRE-RNA is lost. Thus, when iron concentrations increase and iron–sulfur cluster synthesis increases,55 IRP1 conversion to cytoplasmic aconitase increases and IRP1 available to bind to IRE-RNA decreases. IRP2, which does not bind an iron sulfur cluster requires a more complex set of iron-dependent reactions to control. IRP2 is degraded when cellular iron concentrations increase because iron activates FBXL5, a protein that specifically attaches ubiquitin to IRP2 and initiates a protein degradation cascade; living cells degrade the modified IRP2 in organelles called proteosomes. Thus, increased cellular concentrations of iron have three effects that minimize IRE-RNA–IRP binding: (1) inactivation of IRP1 by insertions of an Fe–S cluster into the RNA binding site.9 (2) Iron-dependent modifying enzymes (E3 ligase) that specifically enhance IRP2 degradation by normal proteosomes56,57 and (3) changing IRE-RNA conformations to weaken IRP-binding.11
VII. Mg2+ and Fe2+ influence IRE-RNA binding to EIF4F
The IRE riboregulator binds eIF4F, a protein that contains a ribosome binding site and enhances translation of all mRNAs.51 After an IRE-RNA binds eIF4F, allowing assembly of the mRNA–ribosome complex, a special initiator t-RNA, called met t-RNAi, binds downstream of the IRE structure (toward the 3′ end of the mRNA) allowing protein biosynthesis (mRNA translation) to begin. In the case of IRE-mRNAs, however, both a general and a specific protein interaction is involved. Both the IRE-specific translation repressor protein, IRP, and the generic translation enhancer protein EIF-4F bind to the IRE-RNA structure and compete with each other for IRE-RNA binding;10,11,58 IRP binding prevents IRE-mRNA–ribosome binding and eIF4F facilitates IRE-mRNA–ribosome binding.EIF4-F binding to the IRE-mRNA is enhanced11 by Fe2+, the metabolic regulator of ferritin biosynthesis and other IRE-mRNA encoded proteins of iron homeostasis in animals. Both the equilibrium and kinetics of eIF4F binding to IRE-mRNA are controlled by metal ions and allow eIF-4F binding to outcompete IRP binding. For example, in the absence of metal ions, eIF-4F binding was five times slower than IRP favoring repression of the synthesis of proteins encoded in IRE-mRNAs. IRP–IRE-RNA complexes also have a shorter half-life than eIF-4F/IRE-RNA. However using Mn2+ as an air-resistant proxy for Fe2+, the Kon for eI4F/IRE-RNA complexes increased three fold.46 Thus, both the stability and the binding kinetics of IRE-RNA for the IRP repressor and for binding of the eIF-4F enhancer combine to create metal-dependent increases in IRE-mRNA–ribosome interactions and the synthesis rates of proteins encoded in IRE-mRNAs. While the sites for IRE-RNA conformational change induced by IRP binding, are known (C8 and G16),59,60 as well as for conformational changes induced by metal ions (measured by 2-aminopurine fluorescence to be C8 and A15)35 the specific IRE-RNA binding site(s) for Fe2+ are not known, to date. Hydroxyl radical “hot spots”, generated by Fe2+/H2O2 in air, are known by the induced cleavage “hot spots”, which indirectly indicate proximity to bound Fe2+; they are at the ferritin IRE-RNA hairpin loop residues G6, C7, U8,9 and C23, G24 G25, opposite each other in RNA double helix. The putative Fe2+ sites are in the conserved internal loop bulge characteristic of IRE-RNA stem loops.47 Direct observation of Fe2+ at the ferritin IRE-RNA internal loop, following the sites suggested by hydroxyl radical footprinting with Fe2+ generated radicals,47 using an Mn2+ proxy and NMR spectroscopy, for example, remains a study for investigators of the future.
VIII. Perspective
Iron sensitive mRNAs (IRE-mRNAs) directly bind Fe2+, and encode a group of proteins that control iron balance in animals. Selective metal–RNA interactions occur in RNAs besides IRE-mRNAs, exemplified by ribozymes.61 When the IRE is in the 5′-untranslated region of an mRNA, and this near the ribosome binding site,62 Fe2+-IRE binding changes the RNA conformation to increase synthesis of proteins for managing iron in two ways:(1) Binding of an IRE-specific protein IRP is inhibited.
(2) Binding of a generic protein synthesis enhancer protein, eIF-4F is facilitated.
Riboregulator families, such as the IRE-RNA family, are currently rare; other types of mRNA regulation such as regulated mRNA turnover, and some riboswitches, are more common. Identification rests very high sequence conservation, small size to insure single exon location, and the heavy dependence of bioinformatics tools on linear information. More such RNA families will likely be found with search tools that recognize higher order RNA structure. IRE-RNA is a stem loop, CAGUCX, which contains a trans-loop C–G base pair that creates a tri-loop, AGU. In the base-paired stem of IRE-RNA, the bent RNA A helix contains an unpaired C. Both the terminal triloop and bulge C are contact points for the IRP repressor… However, unlike the hairpin loop and the bulge C, the composition of the IRE helix base pairs is specific for each mRNA in the IRE family and creates an array of different IRP–RNA binding stabilities. As a result, the effect of Fe2+ IRE RNA complexation on protein biosynthesis rates is quantitatively different. Thus, when free iron concentrations increase in cells, ferritin protein synthesis rates increases more than the housekeeping protein, aconitase, because when iron concentrations were low, a larger fraction of ferritin IRE mRNA molecules were inactivated by IRP binding than aconitase IRE-RNA molecules.
The Fe2+ biological signal creates a regulatory feedback loop where the Fe2+ signal is consumed by the protein synthesis product, ferritin. As a result, ferritin protein lowers the free iron concentration, increases IRP binding to ferritin mRNA and decreases ferritin protein synthesis rates. Metal–RNA regulatory reactions illustrated by the shapely, noncoding IRE-RNA family, illustrate the sophistication Nature can employ to rapidly modulate gene activity in the cytoplasm while protecting the master DNA structure in the vault of the nucleus. The effectiveness of hierarchal (quantitatively varied), metal–mRNA–protein interactions, illustrated by the Fe2+–IRE-/IRP family, suggest that the rarity of our current knowledge of such interactions is only temporal and that many more such regulatory families remain to be discovered in the future.
References
- F. Jacob and J. Monod, J. Mol. Biol., 1961, 3, 318–356 CrossRef CAS.
- E. Bonneau and P. Legault, Biochemistry, 2014, 53, 579–590 CrossRef CAS PubMed.
- K. Zeth, Biochem. J., 2012, 445, 297–311 CrossRef CAS PubMed.
- D. Lai, J. R. Proctor and I. M. Meyer, RNA, 2013, 19, 1461–1473 CrossRef CAS PubMed.
- E. J. White, G. Brewer and G. M. Wilson, Biochim. Biophys. Acta, 2013, 1829, 680–688 CrossRef CAS PubMed.
- R. Erlitzki, J. C. Long and E. C. Theil, J. Biol. Chem., 2002, 277, 42579–42587 CrossRef CAS PubMed.
- P. Piccinelli and T. Samuelsson, RNA, 2007, 13, 952–966 CrossRef CAS PubMed.
- S. van Zalen, G. R. Jeschke, E. O. Hexner and J. E. Russell, Blood, 2012, 119, 1045–1053 CrossRef CAS PubMed.
- W. E. Walden, A. I. Selezneva, J. Dupuy, A. Volbeda, J. C. Fontecilla-Camps, E. C. Theil and K. Volz, Science, 2006, 314, 1903–1908 CrossRef CAS PubMed.
- D. J. Goss and E. C. Theil, Acc. Chem. Res., 2011, 44, 1320–1328 CrossRef CAS PubMed.
- J. Ma, S. Haldar, M. A. Khan, S. D. Sharma, W. C. Merrick, E. C. Theil and D. J. Goss, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 8417–8422 CrossRef CAS PubMed.
- S. A. Anderson, C. P. Nizzi, Y. I. Chang, K. M. Deck, P. J. Schmidt, B. Galy, A. Damnernsawad, A. T. Broman, C. Kendziorski, M. W. Hentze, M. D. Fleming, J. Zhang and R. S. Eisenstein, Cell Metab., 2013, 17, 282–290 CrossRef CAS PubMed.
- A. T. McKie, P. Marciani, A. Rolfs, K. Brennan, K. Wehr, D. Barrow, S. Miret, A. Bomford, T. J. Peters, F. Farzaneh, M. A. Hediger, M. W. Hentze and R. J. Simpson, Mol. Cell, 2000, 5, 299–309 CrossRef CAS.
- D. M. Ward and J. Kaplan, Biochim. Biophys. Acta, 2012, 1823, 1426–1433 CrossRef CAS PubMed.
- D. Proudhon, J. Wei, J. Briat and E. C. Theil, Mol. Evol., 1996, 42, 325–336 CrossRef CAS.
- M. Ragland, J. F. Briat, J. Gagnon, J. P. Laulhere, O. Massenet and E. C. Theil, J. Biol. Chem., 1990, 265, 18339–18344 CAS.
- G. Vigani, G. Zocchi, K. Bashir, K. Philippar and J. F. Briat, Trends Plant Sci., 2013, 18, 305–311 CrossRef CAS PubMed.
- W. Wang, M. A. Knovich, L. G. Coffman, F. M. Torti and S. V. Torti, Biochim. Biophys. Acta, 2010, 1800, 760–769 CrossRef CAS PubMed.
- A. A. Alkhateeb and J. R. Connor, Biochim. Biophys. Acta, 2013, 1836, 245–254 CAS.
- K. J. Hintze, Y. Katoh, K. Igarashi and E. C. Theil, J. Biol. Chem., 2007, 282, 34365–34371 CrossRef CAS PubMed.
- K. Igarashi and M. Watanabe-Matsui, Tohoku J. Exp. Med., 2014, 232, 229–253 CrossRef CAS PubMed.
- D. J. Dix, P. N. Lin, Y. Kimata and E. C. Theil, Biochemistry, 1992, 31, 2818–2822 CrossRef CAS.
- Y. Kimata and E. C. Theil, Plant Physiol., 1994, 104, 263–270 CAS.
- K. Hailemariam, K. Iwasaki, B. W. Huang, K. Sakamoto and Y. Tsuji, J. Cell Sci., 2010, 123, 3863–3871 CrossRef CAS PubMed.
- J. D. Webb, M. L. Coleman and C. W. Pugh, Cell. Mol. Life Sci., 2009, 66, 3539–3554 CrossRef CAS PubMed.
- M. Mariotti, P. G. Ridge, Y. Zhang, A. V. Lobanov, T. H. Pringle, R. Guigo, D. L. Hatfield and V. N. Gladyshev, PLoS One, 2013, 7, e33066 Search PubMed.
- D. Fajardo, B. Schlautman, S. Steffan, J. Polashock, N. Vorsa and J. Zalapa, Gene, 2014, 536, 336–343 CrossRef CAS PubMed.
- S. V. Novoselov, M. Rao, N. V. Onoshko, H. Zhi, G. V. Kryukov, Y. Xiang, D. P. Weeks, D. L. Hatfield and V. N. Gladyshev, EMBO J., 2002, 21, 3681–3693 CrossRef CAS PubMed.
- S. S. Merchant, S. E. Prochnik, O. Vallon, E. H. Harris, S. J. Karpowicz, G. B. Witman, A. Terry, A. Salamov, L. K. Fritz-Laylin, L. Marechal-Drouard, W. F. Marshall, L. H. Qu, D. R. Nelson, A. A. Sanderfoot, M. H. Spalding, V. V. Kapitonov, Q. Ren, P. Ferris, E. Lindquist, H. Shapiro, S. M. Lucas, J. Grimwood, J. Schmutz, P. Cardol, H. Cerutti, G. Chanfreau, C. L. Chen, V. Cognat, M. T. Croft, R. Dent, S. Dutcher, E. Fernandez, H. Fukuzawa, D. Gonzalez-Ballester, D. Gonzalez-Halphen, A. Hallmann, M. Hanikenne, M. Hippler, W. Inwood, K. Jabbari, M. Kalanon, R. Kuras, P. A. Lefebvre, S. D. Lemaire, A. V. Lobanov, M. Lohr, A. Manuell, I. Meier, L. Mets, M. Mittag, T. Mittelmeier, J. V. Moroney, J. Moseley, C. Napoli, A. M. Nedelcu, K. Niyogi, S. V. Novoselov, I. T. Paulsen, G. Pazour, S. Purton, J. P. Ral, D. M. Riano-Pachon, W. Riekhof, L. Rymarquis, M. Schroda, D. Stern, J. Umen, R. Willows, N. Wilson, S. L. Zimmer, J. Allmer, J. Balk, K. Bisova, C. J. Chen, M. Elias, K. Gendler, C. Hauser, M. R. Lamb, H. Ledford, J. C. Long, J. Minagawa, M. D. Page, J. Pan, W. Pootakham, S. Roje, A. Rose, E. Stahlberg, A. M. Terauchi, P. Yang, S. Ball, C. Bowler, C. L. Dieckmann, V. N. Gladyshev, P. Green, R. Jorgensen, S. Mayfield, B. Mueller-Roeber, S. Rajamani, R. T. Sayre, P. Brokstein, I. Dubchak, D. Goodstein, L. Hornick, Y. W. Huang, J. Jhaveri, Y. Luo, D. Martinez, W. C. Ngau, B. Otillar, A. Poliakov, A. Porter, L. Szajkowski, G. Werner, K. Zhou, I. V. Grigoriev, D. S. Rokhsar and A. R. Grossman, Science, 2007, 318, 245–250 CrossRef CAS PubMed.
- E. A. Leibold and H. N. Munro, J. Biol. Chem., 1987, 262, 7335–7341 CAS.
- M. W. Hentze, T. A. Rouault, S. W. Caughman, A. Dancis, J. B. Harford and R. D. Klausner, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 6730–6734 CrossRef CAS.
- E. C. Theil and R. S. Eisenstein, J. Biol. Chem., 2000, 275, 40659–40662 CrossRef CAS PubMed.
- D. M. Mauger, N. A. Siegfried and K. M. Weeks, FEBS Lett., 2013, 587, 1180–1188 CrossRef CAS PubMed.
- I. M. Silverman, F. Li and B. D. Gregory, Plant Sci., 2013, 205–206, 55–62 CrossRef CAS PubMed.
- M. A. Khan, W. E. Walden, D. J. Goss and E. C. Theil, J. Biol. Chem., 2009, 284, 30122–30128 CrossRef CAS PubMed.
- Z. Gdaniec, H. Sierzputowska-Gracz and E. C. Theil, Biochemistry, 1999, 38, 5676 CrossRef PubMed.
- S. A. McCallum and A. Pardi, J. Mol. Biol., 2003, 326, 1037–1050 CrossRef CAS.
- E. C. Theil and D. J. Goss, Chem. Rev., 2009, 109, 4568–4579 CrossRef CAS PubMed.
- J. B. Goforth, S. A. Anderson, C. P. Nizzi and R. S. Eisenstein, RNA, 2010, 16, 154–169 CrossRef CAS PubMed.
- Y. Ke, J. Wu, E. A. Leibold, W. E. Walden and E. C. Theil, J. Biol. Chem., 1998, 273, 23637–23640 CrossRef CAS PubMed.
- K. L. Schalinske, O. S. Chen and R. S. Eisenstein, J. Biol. Chem., 1998, 273, 3740–3746 CrossRef CAS PubMed.
- C. O. dos Santos, L. C. Dore, E. Valentine, S. G. Shelat, R. C. Hardison, M. Ghosh, W. Wang, R. S. Eisenstein, F. F. Costa and M. J. Weiss, J. Biol. Chem., 2008, 283, 26956–26964 CrossRef CAS PubMed.
- K. L. Schalinske, O. S. Chen and R. S. Eisenstein, J. Biol. Chem., 1998, 273, 3740–3746 CrossRef CAS PubMed.
- T. M. Schmeing, K. S. Huang, D. E. Kitchen, S. A. Strobel and T. A. Steitz, Mol. Cell, 2005, 20, 437–448 CrossRef CAS PubMed.
- P. Auffinger, N. Grover and E. Westhof, Met. Ions Life Sci., 2011, 9, 1–35 CAS.
- M. A. Khan, J. Ma, W. E. Walden, W. C. Merrick, E. C. Theil and D. J. Goss, Nucleic Acids Res., 2014, 42, 6567–6577 CrossRef CAS PubMed.
- C. M. Harrell, A. R. McKenzie, M. M. Patino, W. E. Walden and E. C. Theil, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 4166–4170 CrossRef CAS.
- D. Moazed, S. Stern and H. F. Noller, J. Mol. Biol., 1986, 187, 399–416 CrossRef CAS.
- S. Mohammed, M. M. Phelan, U. Rasul and V. Ramesh, Org. Biomol. Chem., 2014, 12, 1495–1509 CAS.
- J. D. Tibodeau, P. M. Fox, P. A. Ropp, E. C. Theil and H. H. Thorp, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 253–257 CrossRef CAS PubMed.
- C. C. Thoreen, Biochem. Soc. Trans., 2013, 41, 913–916 CrossRef CAS PubMed.
- T. Lee and J. Pelletier, Future Med. Chem., 2012, 4, 19–31 CrossRef CAS PubMed.
- J. Zahringer, B. S. Baliga and H. N. Munro, Proc. Natl. Acad. Sci. U. S. A., 1976, 73, 857–861 CrossRef CAS.
- L. F. Dickey, Y. H. Wang, G. E. Shull, I. A. Wortman 3rd and E. C. Theil, J. Biol. Chem., 1988, 263, 3071–3074 CAS.
- D. J. Netz, J. Mascarenhas, O. Stehling, A. J. Pierik and R. Lill, Trends Cell Biol., 2014, 24, 303–312 CrossRef CAS PubMed.
- A. A. Salahudeen, J. W. Thompson, H. W. Ma, L. N. Kinch, Q. Li, N. V. Grishin and R. K. Bruick, Science, 2009, 326, 722–726 CrossRef CAS PubMed.
- K. M. Deck, A. Vasanthakumar, S. A. Anderson, J. B. Goforth, M. C. Kennedy, W. E. Antholine and R. S. Eisenstein, J. Biol. Chem., 2009, 284, 12701–12709 CrossRef CAS PubMed.
- M. A. Khan and D. J. Goss, Biochemistry, 2005, 44, 4510–4516 CrossRef CAS PubMed.
- W. E. Walden, A. I. Selezneva, J. Dupuy, A. Volbeda, J. C. Fontecilla-Camps, E. C. Theil and K. Volz, Science, 2006, 314, 1903–1908 CrossRef CAS PubMed.
- A. I. Selezneva, W. E. Walden and K. W. Volz, J. Mol. Biol., 2013, 425, 3301–3310 CrossRef CAS PubMed.
- D. M. Truong, D. J. Sidote, R. Russell and A. M. Lambowitz, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E3800–E3809 CrossRef CAS PubMed.
- M. L. Wallander, E. A. Leibold and R. S. Eisenstein, Biochim. Biophys. Acta, 2006, 1763, 668–689 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |