IRE mRNA riboregulators use metabolic iron (Fe2+) to control mRNA activity and iron chemistry in animals
Abstract
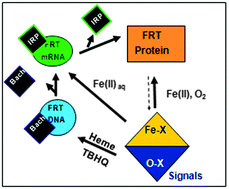
A family of noncoding RNAs bind Fe2+ to increase protein synthesis. The structures occur in messenger RNAs encoding animal proteins for iron metabolism. Each mRNA regulatory sequence, ∼30 ribonucleotides long, is called an IRE (Iron Responsive Element), and folds into a bent, A-RNA helix with a terminal loop. Riboregulatory RNAs, like t-RNAs, r-RNAs micro-RNAs, etc. contrast with DNA, since single-stranded RNA can fold into a variety of complex, three-dimensional structures. IRE-RNAs bind two types of proteins: (1) IRPs which are protein repressors, sequence-related to mitochondrial aconitases. (2) eIF-4F, which bind ribosomes and enhances general protein biosynthesis. The competition between IRP and eIF-4F binding to IRE-RNA is controlled by Fe2+-induced changes in the IRE-RNA conformation. Mn2+, which also binds to IRE-RNA in solution, is a convenient experimental proxy for air-sensitive Fe2+ studies of in vitro protein biosynthesis and protein binding. However, only Fe2+ has physiological effects on protein biosynthesis directed by IRE-mRNAs. The structures of the IRE-RNA riboregulators is known indirectly from effects of base substitutions on function, from solution NMR of the free RNA, and of X-ray crystallography of the IRE-RNA–IRP repressor complex. However, the inability to date, to crystallize the free IRE-RNA, and the dissociation of the IRE-RNA–IRP complex when metal binds, have hampered direct identification and characterization of the RNA–metal binding sites. The high conservation of the primary sequence in IRE-mRNA control elements has facilitated their identification and analysis of metal-assisted riboregulator function. Expansion of RNA search analyses beyond primary will likely reveal other, metal-dependent families of mRNA riboregulators.

 Please wait while we load your content...
Please wait while we load your content...