Virtual screening for novel Atg5–Atg16 complex inhibitors for autophagy modulation†
Elizabeth Robinson a, Euphemia Leung b, Anna M. Matuszek c, Niels Krogsgaard-Larsen c, Daniel P. Furkert c, Margaret A. Brimble c, Alan Richardson a and Jóhannes Reynisson *c
aInstitute for Science and Technology in Medicine, Keele University, Guy Hilton Research Centre, Stoke-on-Trent, UK
bAuckland Cancer Society Research Centre, University of Auckland, New Zealand
cSchool of Chemical Sciences, University of Auckland, Private Bag 92019, Auckland 1142, New Zealand. E-mail: j.reynisson@auckland.ac.nz; Fax: +64-9-373-7422; Tel: +64-9-373-7599 ext. 83746
First published on 3rd November 2014
Abstract
Two hit compounds (14 and 62) were identified using virtual high throughput screening (vHTS) inhibiting the autophagy process in A2780 ovarian cancer cells. The expression levels of the LC3-II and p62 autophagy marker proteins were monitored using Western blotting. Preliminary structure activity relationship (SAR) study of close structural analogues revealed another active compound 38. The three active compounds were tested in the MCF-7 human breast cancer cells and severe reduction of autophagosomes formation was observed confirming the activity of the inhibitors. The docking scaffold used for the vHTS was a lipophilic cleft on the Atg5 protein, which is occupied by a phenylalanine residue in the Atg16 polypeptide. To the best of our knowledge this is the first report on inhibitors that specifically modulate autophagy by directly inhibiting autophagy specific proteins, which is significant due the role autophagy plays in a number of morbid diseases such as cancer.
Introduction
Autophagy (self-eating) is a catabolic pathway that sequesters undesired cellular material into autophagosomes for delivery to lysosomes for the bulk degradation and is conserved in most eukaryotes.1 Autophagic dysfunction is now associated with a number of diseases, including infection by pathogens, inflammatory bowel disease, neurodegeneration and cancer.2–4 A number of small molecules are known to modulate autophagy however there is a dearth of compounds that directly interact with the specific autophagy proteins.5 A key step in the pathway is the covalent conjugation of the ubiquitin-related protein Atg8 (autophagy) to phosphatidylethanolamine in autophagic membranes by a complex consisting of Atg16 and the Atg12–Atg5 conjugate.1 The crystal structure of the Atg5–Atg16 complex is known and in vitro analysis showed that the Arg-35 and Phe-46 amino acids of Atg-16 are crucial for this interaction to take place.6,7 The binding pockets are shown in Fig. 1. This protein complex therefore represents a new target for drug development,8 in particular for a virtual high throughput screening (vHTS) campaign, which is an effective method to generate inhibitors of bio-molecular targets.9–11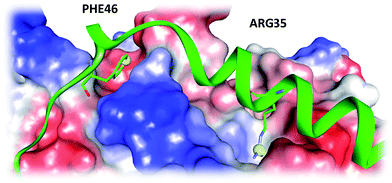 | ||
| Fig. 1 The two binding pockets of interest of the Atg5–Atg16 complex identified by mutation studies. The Atg5 protein surface is rendered and the Atg16 polypeptide is green. The amino acids Arg35 and Phe46 are shown in the stick format. | ||
In this study, the crystal structure of the Atg5–Atg16 complex at the binding site of Phe-46 in Atg-16 was used as a docking scaffold for a vHTS to find small molecular inhibitors for the autophagy process in ovarian and breast cancer cell lines.
Results
Pilot screen
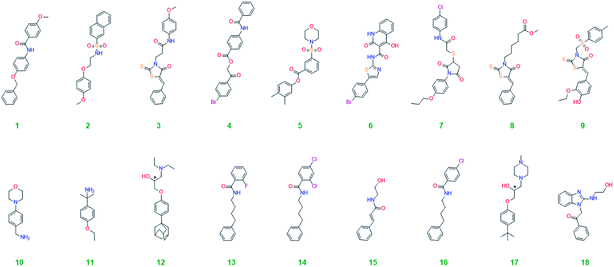
1 × 104 molecular entities were downloaded from the ChemBridge diversity collection.12 It was filtered to render the list Lipinski13 compliant, resulting in 8373 molecules. These were screened against the cleft in Atg5 which the phenyl moiety of Phe-46 from the Atg16 polypeptide occupies. The binding pocket at Arg-35 (see Fig. 1) was also considered but several amino acidic were missing from the crystal structure, which are a part of a flexible loop directly in this pocket. The proximity of the loop makes the shape of the pocket uncertain and less attractive as a docking scaffold than the cleft for Phe-46 and it was therefore not considered further. The GoldScore14 and ChemScore15,16 fitness functions were used for the initial screen. All ligands without predicted hydrogen bonding were eliminated as well as those with low ChemScore (<10); no such low values were predicted by GoldScore. This left 512 candidates, which were screened again with high search efficiency using both scoring functions. The candidates were eliminated based on the GoldScore (≤30) and ChemScore (≤20); and the ability to form hydrogen bonds based on both scoring functions resulting in 33 candidates. The results were inspected visually for consensus of the best predicted configuration of the ligands between the scoring functions, that the ligands had plausible configurations, i.e., not strained, for lipophilic moieties pointing into the water environment and, finally for undesirable moieties linked to cell toxicity and chemical reactivity.17 The screening methodology used has been previously successfully applied to find active ligands of the phosphoinositide specific-phospholipase C-γ2 enzyme.18 Eighteen compounds were selected for experimental testing and their structures are shown in Table 1. A detailed description of the vHTS is given in the Methodology section.Biological testing
Microtubule-associated protein light chain 3 (LC3-II) is the mammalian ortholog of yeast Atg-8 and a robust marker of autophagosomes.19 LC3 binds the polyubiquitin-binding protein p62, an adapter protein that is selectively degraded via autophagy.8 The eighteen virtual hits were tested in A2780 ovarian tumour cell line for efficacy. Autophagy was induced by starvation (PBS buffer for 3 hours) and the effect of the ligands was assessed by measuring accumulation of LC3-II and p62 by immunoblotting (see methodology for detailed description). A decrease in LC3-II and increase in p62 is consistent with suppression of autophagy. Two set of experiments were conducted for each compound. The representative results are shown in Fig. 2 for compounds 10–18 and in Fig. S1† in the ESI for compounds 1–9; quantitation of both sets of data is also provided in the ESI (Fig. S6 and S7†).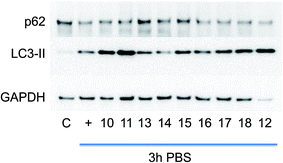 | ||
| Fig. 2 Immunoblotting of p62 and LC3-II protein in ovarian cancer A2780 cell line. Cells were incubated with DMSO as negative control (C), or PBS for 3 h to induce autophagy without (+) or with pre-incubation of compounds for 1 h. p62 and LC3-II were detected in total cell extracts by western blotting analysis, using specific antibodies. GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was used as loading control. The quantitative results are given in Fig. S7 in the ESI.† | ||
The control profile of the cellular proteins as base line was shown on the left lane in Fig. 2. Upon induction of autophagy, the expected decrease in LC3-II and increase in p62 was observed. Compounds 13 and 14 reduced accumulation of LC3-II and depletion of p62. Dose response experiments were performed to verify this activity of these ligands (Fig. S2 and S3 in the ESI section†). These experiments demonstrated the activity of compound 14 was dose dependent (Fig. 3), but this was not apparent for ligand 13.
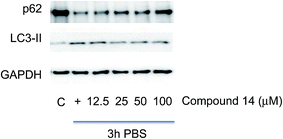 | ||
| Fig. 3 Dose response effect for ligand 14 using immunoblotting of p62 and LC3-II protein in ovarian cancer A2780 cell line. Cells were incubated with DMSO as negative control (C), or PBS for 3 h to induce autophagy (+) without or with pre-incubation of indicated concentration of compound 14 for 1 h. GAPDH was used as loading control. The quantitative result for compound 14 is given in Fig. S8 in the ESI.† | ||
It can be clearly seen in Fig. 3 that an increased concentration of ligand 14 causes the re-emergence of p62 compared to the second lane (+) of autophagy cells and the reduction of LC3-II, which is a strong indication that the ovarian cell are reverted from their autophagy state.
Preliminary structural activity relationship (SAR)
In order to explore the chemical space around the hit compound 14 ten close structural analogues were purchased and tested. In addition two compounds (13 and 16) from the pilot screen are similar to 14 resulting in thirteen molecules for the SAR analysis (Scheme 1).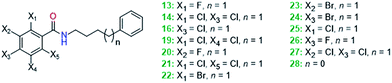 | ||
| Scheme 1 The structures of twelve close structural analogues of hit compound 14, which were tested for their activity for SAR generation. | ||
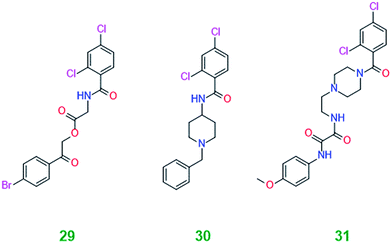
Various single and double chlorine substitutions of the benzamide ring did not lead to active compounds suggesting that the ortho/para substitution pattern is crucial for the efficacy observed. Ortho and para fluorine single substitutions (13 and 26) on the benzamide ring were not tolerated. Furthermore, bromine single substitution (ortho (22), meta (23) and para (24)) gave inconsistent effects on the expression levels of LC3-II and p62. There is not a clear indication of inhibition and it can be concluded that single bromine substitution of the benzamide ring is not favourable. Finally, one derivative (28) with a shorter aliphatic chain (n = 0) and no substitution on the ring systems was not active. In conclusion, the ortho/para chlorine substitution pattern is vital for the activity. Three compounds, 29–31, with the ortho/para chlorine substitution on the benzamide were tested and their structures are shown in Table 2. None of the ligands demonstrated consistent activity, which suggests that the dichlorinated benzamide moiety is on its own insufficient to modulate autophagy.
Synthetic chemistry
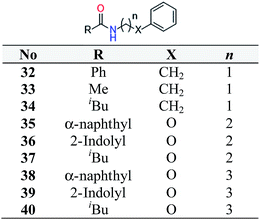
In order to expand the preliminary SAR further nine more analogues were synthesised of hit compound 14 (Scheme 2). Three features were deemed important to investigate: first, the tolerance of the benzamide phenyl moiety for substitution with other functional groups; secondly the length of the alkyl chain and thirdly the tolerance of oxygen substitution in the alkyl chain.Biological testing revealed that only compound 38 showed activity, giving increased p62 and decreased LC3-II expression. This was verified with a dose response experiment. The efficacy of ligand 38 demonstrates that the substitution of the benzamide phenyl moiety with α-naphthyl is possible and that introduction of an ether link into the alkyl chain is also tolerated. The length of the linker chain is clearly important, with four units optimal, as seen for both hit compound 14 and its derivative 38 compared to ligands 32–37, which have a shorter chain length. Even though twenty five compounds were tested in this preliminary analysis, a clear SAR was not evident. Thus, substantially more analogues of the hit compounds need to be tested in order to produce a full SAR.
Main screen
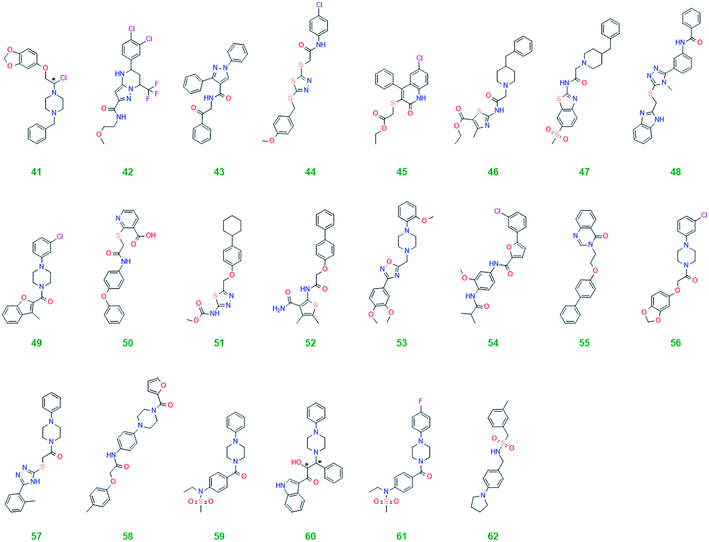
For the main screen, the complete diversity ChemBridge diversity collection was used consisting of 5 × 104 molecules.12 After making it Lipinski compliant 4.3 × 104 entities remained. All four scoring functions available in the GOLD software suite (GoldScore,14 ChemScore,14,15 ChemPLP20 and ASP21) were used in conjunction with the consensus concept, i.e., only ligands that scored well with all the functions and showed hydrogen bonding activity were taken forward.22,23 For the initial screen it sufficed to take only compounds with good hydrogen bonding (HB ≥ 1) forward resulting in 1470 candidates. For the second screen, using higher search efficiency and more docking runs, again only ligands with good hydrogen bonding were taken forward as well as a good score (GoldFitness ≥ 45, ChemSocre ≥ 20, PLPScore ≥ 50 and ASP ≥ 25) leaving 200 candidates. These were visually inspected using the same criteria as for the pilot screen. Twenty-two ligands were selected for experimental testing and their structures are shown in Table 3.From the twenty-two virtual hits tested in the ovarian cancer cells ligand 62 caused an increase in p62 and decrease in LC3-II consistent with inhibition of autophagy (see Fig. S4†). This was also seen for ligand 58 to some extent but the effect was less pronounced than for 62.
Hit verification
In order to verify the activity of the three hit compounds 14, 38 and 62 in blocking autophagy cells back to the normal state they were tested in the MCF-7 human breast cancer cell line. The results are shown in Fig. 4.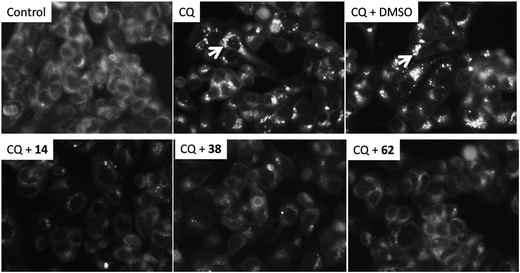 | ||
| Fig. 4 The effect of pre-treating MCF-7 human breast cancer cells with hit compounds 14, 38 and 62 before incubation with 75 μM chloroquine (CQ). Autophagolysosomes were visualized by monodansylcadaverine (MDC) staining (see arrows). MDC was loaded into cells and incubated at 37 °C for 10 min and visualized by fluorescent microscopy. The pre-incubation with the hit compounds abrogated the CQ induced autophagolysosomes accumulation. | ||
Chloroquine (CQ) is a lysosomotropic agent that prevents autophagy protein degradation by inhibiting lysosome-mediated proteolysis and causes autophagolysosomes accumulation.24 The auto-fluorescent compound monodansylcadaverine (MDC) is a widely used marker of autophagolysosomes as it labels vacuoles formation in later stages in the autophagy degradation process.25–27 Incubation of cells with 75 μM CQ caused a significant increase in the number of autophagolysosomes, visualized by MDC accumulation in vacuoles (Fig. 4). The administration of the hits, 14, 38 and 62, resulted in effective suppression of the autophagolysosome formation in line with the hypothesis of Atg5–Atg16 complex inhibition. Finally, hit 62 was tested in the well-established National Cancer Institute's NCI60 panel of 60 human tumour cells lines.28 It had a marginal effect on the growth of the cell lines with a mean of 90.1% at 10 μM as compared to 100% of untreated cells. These data suggest that the effect of compound 62 was not dependent on inhibition of cell growth. The result from the NCI60 experiment is shown in Fig. S5 in the ESI.†
Modelling of hit compounds
The binding to Atg5 of the three hit compounds was investigated. The predicted binding modes of hit 14 are shown in Fig. 5.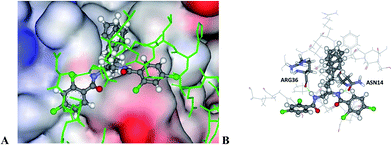 | ||
| Fig. 5 The docked configuration of 14 in the binding site of Atg5 using GoldScore and ChemScore. (A) The protein surface is rendered. The phenyl group of 14 occupies a lipophilic cleft overlapping with the Phe46 amino acid in Atg16, which is shown as green sticks. Red depicts a positive partial charge on the surface, blue depicts negative partial charge and grey shows neutral/lipophilic areas. (B) Hydrogen bonds are shown as green lines between ligand 14 and the amino acids Arg36 for the GoldScore results and Asn14 for the ChemScore configuration. | ||
The modelling of 14 resulted in two plausible binding modes to Atg5. GoldScore and ChemPLP predict the same conformation with hydrogen bonding to the backbone carboxylic moiety of Arg36 but ChemScore predicts bonding to the backbone carboxylic moiety of Asn14 (Fig. 5B). In both cases the phenyl moiety of 14 occupies the lipophilic cleft in place of Phe46 of the Atg16 polypeptide and overlaps with it in general (Fig. 5A). The ASP scoring function produced a different binding mode to its counterparts but the lipophilic cleft was also occupied by the phenyl moiety. Hit compounds 38 and 62 gave less consistent results when modelled but in all cases the lipophilic binding cleft was filled, suggesting that this may be necessary for the efficacy of the compounds.
Discussion
It is becoming more apparent that the autophagy process plays a pivotal role in diseases such as cancer, e.g., apoptotic cancer cells induced by chemotherapeutics can not only survive but recover from damage with the aid of heightened autophagy activity.29 The implication is that inhibiting autophagy with small molecules may increase the efficacy of chemotherapeutic regimes.The three hits found in this project are a good start in developing effective inhibitors for autophagy. At this stage, we can not rule out the possibility that the compounds inhibit autophagy through some mechanism other than interfering with Atg5–Atg16 complex formation. The development of a specific biochemical assay, isothermal calorimetric titration (ICT) experiments and/or X-ray crystallography will answer this vital question. In particular, the results of ICT experiments can reveal whether the hits can perturb the Atg5–Atg16 complex formation and we are currently evaluating this. Furthermore, the exploration of chemical space surrounding the hits by chemical synthesis will produce a more complete SAR, identifying more potent ligands and aiding in developing a better molecular model. The SAR study presented here can only be viewed as preliminary. Nevertheless, having the three hit compounds identified in this work is the crucial start to give traction to develop autophagy-specific inhibitors and hopefully leading to an effective adjunctive therapeutic approach in the treatment of cancer.
Conclusion
In this work two hit compounds 14 and 62 were identified using virtual high throughput screening inhibiting autophagy in ovarian A2780 cancer cells. Preliminary SAR was developed for hit 14 leading to the discovery of another active compound 38. The three active compounds were tested in MCF-7 breast cancer cells and the characteristic autophagy vacuole formation was diminished upon the administration of the inhibitors verifying their efficacy. These three compounds can now be developed further with more elaborate experimental/biological testing and by substantially extending the SAR analysis.Methodology
Modelling and screening
The compounds were docked to the crystal structure of Atg16–Atg5 (PDB ID: 2DYO, resolution 1.97 Å),6 which was obtained from the Protein Data Bank (PDB).30,31 The Scigress Ultra version 7.7.0.47 program32 was used to prepare the crystal structure for docking, i.e., hydrogen atoms were added, the co-crystallised polypeptide Atg-16 was removed as well as crystallographic water molecules. The centre of the binding pocket was defined as the position of the Atg-16's Phe-46 of the para-carbon of the phenyl ring (x = 87.120, y = 88.114, z = 69.571) with 10 Å radius. For the initial screen 30% search efficiency was used (virtual screen) with ten runs per compound. For the second phase (re-dock) 100% efficiency was used in conjunction with fifty docking runs. The GoldScore (GS),14 ChemScore (CS),15,16 ChemPLP20 and ASP21 scoring functions were implemented to validate the predicted binding modes and relative energies of the ligands using the GOLD v5.2 software suite. The virtual high throughput screen was conducted with the ChemBridge diversity collection of 5 × 104 entities.12 Substructure and Tanimoto similarity search methods33 were used to identify the structurally related compounds using the Hit2Lead12 web based compound libraries.Western blotting
The investigational compounds identified through screening for autophagic activity were used in cell-based assays to determine their effects on autophagy in ovarian cancer cells. A2780 cells (5 × 105 cells per well) were plated in 6-well plates in 1 mL of growth medium and pre-incubated for 24 h. Following this, the cells were incubated with 50–100 μM of each compound for 1 h before replacing the medium in each well with PBS supplemented with the corresponding compound. DMSO was added to the control wells to equal the concentration in drug-treated wells (max. 0.5%). After 3 h of PBS-induced autophagy, cells were lysed and autophagy-specific proteins detected and autophagy-specific proteins (LC3-II and p62) detected using western blotting with GAPDH used as loading control. Each compound was tested in two or three independent experiments. Further dose–response assays were performed with investigational compounds that demonstrated autophagy inhibition in the initial assay. Similar assays were performed with four concentrations (12.5 μM, 25 μM, 50 μM and 100 μM) of each compound. The concentration of DMSO was normalised across all wells for each experiment.The buffer used for A2780 ovarian cancer cells lysis was prepared consisting of 20 mM HEPES, 150 mM NaCl, 2 mM EDTA, 0.5% sodium deoxycholate, 1% NP40, 120 μM leupeptin, 1 μM pepstatin and 1 mM PMSF. The concentration of protein within the lysate was established using Sigma supplied bicinchoninic acid kit. Nusep 4–20% polyacrylamide gradient gel was used, loading between 5–15 μg protein per sample. After protein separation on gel, the proteins were transferred onto the PVDF membrane, which was then blocked in 5% skimmed milk powder dissolved in TBST buffer consisting of 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20, for 1 h at RTP. The membrane was then incubated in antibodies for ∼16 h (overnight) at temperature of 4 °C, where LC3-2G6 (Nanotools) at 1/2000 dilution was utilised as anti-LC3 antibody and AB56416 (Abcam) at 1/1000 dilution was used as anti-SQSTM1/p62 antibody. The dilution step was then repeated in 5% milk in TBST as above, and subsequently in anti-GAPDH antibody MAB374 (Millipore) at 1/5000 dilution for 1 h at RTP. After each primary antibody, the membrane was washed three times for 5 min. The membrane was then incubated in anti-mouse IgG antibody linked to HRP (Cell Signalling Technology) at 1/2000 dilution in a room temperature for a period of 1 h. Membrane was then washed in TBST five times consecutively, before the detection of protein bands, which was conducted using UptiLight HRP chemiluminescent substrate (Uptima) and a FluorChem M Imager. To quantitate individual proteins, signal intensity was integrated over the area of each band and corrected for background. Results were normalized to GAPDH measured in the same sample.
MCF-7 breast cancer cells
Human breast cancer cell line MCF-7 grown in 96 well plates were not treated (control) or treated with chloroquine (75 μM) alone or with 14, 38 and 62 at 25 μM. DMSO (0.2%) was incubated for vehicle control. After 5 hours, cells were incubated with 10 μM monodansylcadaverine (MDC, Sigma, 30 432) for 20 min at 37 °C, then washed with PBS three times. Images were examined immediately by fluorescence microscopy (FLoid Cell Imaging Station, 40× magnification).NCI's 60-cell line panel growth inhibition assay
The compounds obtained were submitted to the National Cancer Institute's Developmental Therapeutic Program (DTP) where they were screened against a panel of sixty human tumour cell lines (NCI60,28 for further information see ref. and references therein). The full description of the assay is given in the ESI.†Chemistry
Known compounds 32–34 were prepared by acylation of phenethylamine hydrochloride using standard conditions. Experimental procedures and characterisation data are provided in the ESI.† Compounds 35–40 were prepared from either 2- or 3-phenoxylethylamine, as described below.Acknowledgements
Funding for this work was obtained from the Maurice and Phyllis Paykel trust, the McClelland Trust, Auckland Medical Research Foundation and Genesis Oncology Trust (GOT-1429-PDA). This work is also supported by Auckland Cancer Society Research Centre.References
- A. Kaufmann, V. Beier, H. G. Franquelim and T. Wollert, Cell, 2014, 156, 469–481 CrossRef CAS PubMed.
- N. N. Noda, Y. Ohsumi and F. Inagaki, Chem. Rev., 2009, 109, 1587–1598 CrossRef CAS PubMed.
- E. Morselli, L. Galluzzi, O. Kepp, J.-M. Vicencio, A. Criollo, M. C. Maiuri and G. Kroemer, Biochim. Biophys. Acta, 2009, 1793, 1524–1532 CrossRef CAS PubMed.
- N. B. Nedelsky, P. K. Todd and J. P. Taylor, Biochim. Biophys. Acta, 2008, 1782, 691–699 CrossRef CAS PubMed.
- A. Fleming, T. Noda, T. Yoshimori and D. C. Rubinsztein, Nat. Chem. Biol., 2011, 7, 9–17 CrossRef CAS PubMed.
- M. Matsushita, N. N. Suzuki, K. Obara, Y. Fujioka, Y. Ohsumi and F. Inagaki, J. Biol. Chem., 2007, 282, 6763–6772 CrossRef CAS PubMed.
- Y. Fujioka, N. N. Noda, H. Nakatogawa, Y. Ohsumi and F. Inagaki, J. Biol. Chem., 2010, 285, 1508–1515 CrossRef CAS PubMed.
- A. Brennand, M. Gualdrón-López, I. Coppens, D. J. Rigden, M. L. Ginger and P. A. M. Michels, Mol. Biochem. Parasitol., 2011, 177, 83–99 CrossRef CAS PubMed.
- R. V. Guido, G. Oliva and A. D. Andricopulo, Curr. Med. Chem., 2008, 15, 37–46 CrossRef CAS.
- T. Zhu, S. Cao, P. C. Su, R. Patel, D. Shah, H. B. Chokshi, R. Szukala, M. E. Johnson and K. E. Hevener, J. Med. Chem., 2013, 12, 6560–6572 CrossRef PubMed.
- T. Scior, A. Bender, G. Tresadern, J. L. Medina-Franco, K. Martínez-Mayorga, T. Langer, K. Cuanalo-Contreras and D. K. Agrafiotis, J. Chem. Inf. Model., 2012, 52, 867–881 CrossRef CAS PubMed.
- Hit2Lead http://www.hit2lead.com/, ChemBridge Corporation.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 1997, 23, 3–25 CrossRef CAS.
- G. Jones, P. Willet, R. C. Glen, A. R. Leach and R. Taylor, J. Mol. Biol., 1997, 267, 727–748 CrossRef CAS PubMed.
- M. D. Eldridge, C. Murray, T. R. Auton, G. V. Paolini and P. M. Mee, J. Comput.-Aided Mol. Des., 1997, 11, 425–445 CrossRef CAS.
- M. L. Verdonk, J. C. Cole, M. J. Hartshorn, C. W. Murray and R. D. Taylor, Proteins, 2003, 52, 609–623 CrossRef CAS PubMed.
- P. Axerio-Cilies, I. P. Castañeda, A. Mirza and J. Reynisson, Eur. J. Med. Chem., 2009, 44, 1128–1134 CrossRef CAS PubMed.
- J. Reynisson, W. Court, C. O'Neill, J. Day, L. Patterson, E. McDonald, P. Workman, M. Katan and S. A. Eccles, Bioorg. Med. Chem., 2009, 17, 3169–3176 CrossRef CAS PubMed.
- K. B. Larsen, T. Lamark, A. Øvervatn, I. Harneshaug, T. Johansen and G. Bjørkøy, Autophagy, 2010, 6, 784–793 CrossRef CAS.
- O. Korb, T. Stützle and T. E. Exner, J. Chem. Inf. Model., 2009, 49, 84–96 CrossRef CAS PubMed.
- W. T. M. Mooij and M. L. Verdonk, Proteins, 2005, 61, 272–287 CrossRef CAS PubMed.
- R. Wang and S. Wang, J. Chem. Inf. Comput. Sci., 2001, 41, 1422–1426 CrossRef CAS PubMed.
- J. M. Yang, Y. F. Chen, T. W. Shen, B. S. Kristal and D. F. Hsu, J. Chem. Inf. Model., 2005, 45, 1134–1146 CrossRef CAS PubMed.
- A. Kawai, H. Uchiyama, S. Takano, N. Nakamura and S. Ohkuma, Autophagy, 2007, 3, 154–157 CrossRef CAS.
- D. J. Klionsky, F. C. Abdalla, H. Abeliovich, R. T. Abraham, A. Acevedo-Arozena and K. Adeli, et al. , Autophagy, 2012, 8, 445–544 CrossRef CAS.
- E. T. Bampton, C. G. Goemans, D. Niranjan, N. Mizushima and A. M. Tolkovsky, Autophagy, 2005, 1, 23–36 CrossRef CAS.
- K. Vanderlaag, Y. Su, A. E. Frankel, R. C. Burghardt, R. Barhoumi, G. Chadalapaka, I. Jutooru and S. Safe, BMC Cancer, 2010, 10, 669 CrossRef CAS PubMed.
- R. H. Shoemaker, Nat. Rev. Drug Discovery, 2006, 6, 813–823 CrossRef CAS PubMed.
- J. Thorburn, Z. Andrysik, L. Staskiewicz, J. Gump, P. Maycotte, A. Oberst, D. R. Green, J. M. Espinosa and A. Thorburn, Cell Rep., 2014, 7, 45–52 CrossRef CAS PubMed.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed.
- H. Berman, K. Henrick and H. Nakamura, Nat. Struct. Biol., 2003, 10, 980 CrossRef CAS PubMed.
- Scigress Explorer Ultra Version 7.7.0.47, Fijitsu Limited, 2000–2007 Search PubMed.
- A. R. Leach and V. J. Gillet, 1. Representation and manipulation of 2D molecular structures, in An Introduction to Chemoinformatics, Kluwer Academic Publisher, Dordrecht, 2003, pp. 1–26 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4md00420e |
| This journal is © The Royal Society of Chemistry 2015 |