Novel isoniazid–amidoether derivatives: synthesis, characterization and antimycobacterial activity evaluation†
Deepak Kumar a, Garima Khare b, Beena a, Saqib Kidwai c, Anil K. Tyagi b, Ramandeep Singh c and Diwan S. Rawat *a
aDepartment of Chemistry, University of Delhi, Delhi-110007, India. E-mail: dsrawat@chemistry.du.ac.in; Tel: +91-11-27667465
bDepartment of Biochemistry, University of Delhi South Campus, New Delhi-110021, India
cTranslational Health Science and Technology Institute, Vaccine and Infectious Disease Research Centre, Gurgaon, 122016, India
First published on 23rd September 2014
Abstract
A series of isoniazid–amidoether derivatives was synthesized and screened for their antimycobacterial activity in vitro and in vivo. Most of the compounds exhibited potent in vitro activity against the Mycobacterium tuberculosis H37Rv strain with MIC99 values ranging from 0.39 to 3.125 μM. Five compounds were equally as potent as the reference compound isoniazid. The most active compound 3b, when evaluated for in vivo activity, exhibited mild reduction in the bacillary load in lungs. However it showed a better effect in spleens. All the compounds were also evaluated for their cytotoxicity against the THP-1 cell line and no toxicity was observed up to 50 μM concentrations. The calculated ADMET parameters for the compounds validated good pharmacokinetic properties, confirming that these compounds could be used as templates for the development of new anti-tuberculosis agents.
Introduction
Tuberculosis (TB) is an infectious disease mainly caused by Mycobacterium tuberculosis. It is a leading cause of death worldwide, infecting about 9.2 million people and killing approximately 2.0 million people annually.1 After the discovery of many effective anti-TB drugs during the 1950s and 1970s, such as ethambutol, isoniazid, pyrazinamide, rifampicin and streptomycin, there was a drastic decline in the number of TB cases, especially in developed countries. However, since the 1980s, the number of TB cases throughout the world has been increasing rapidly due to the emergence of multi-drug resistant tuberculosis (MDR-TB), extensively drug-resistant tuberculosis (XDR-TB) and more recently totally drug resistant tuberculosis (TDR-TB).2 The MDR, XDR and TDR forms of tuberculosis are more often dreadful and difficult to treat.3–7 The TDR-TB strain has been shown to be resistant to all the first line, second line and third line anti-TB drugs. The situation becomes more complicated in human immunodeficiency virus (HIV) infected patients as they are more likely to be infected with TB due to their weak immune system.8,9 However, in recent years several new drug candidates or repurposed drugs namely gatifloxacin, moxifloxacin, rifapentine, TMC207, OPC67683, PA824, linezolid, PNU100480, AZD5847, SQ109, etc. have been developed and some of them are also in the advance stages of clinical trials for the treatment of tuberculosis.10–18 Only one new drug, bedaquiline, has been recently approved by FDA for its use in drug resistant tuberculosis.19–21Due to the global impact of this devastating disease, there is an urgent need for the development of new derivatives with promising antimycobacterial activities. Several different approaches such as targeting bacterial virulence, high-throughput screening (HTS), structure-based drug discovery (SBDD), chemical modifications of the known drugs and combinatorial chemistry have been explored to search novel biologically important molecules.22–24 Among all these strategies, the molecular modification approach has been found to be very promising and several drugs available in the market have been developed by using this strategy. Molecular modification is a chemical change in a molecule with the aim to enhance its pharmaceutical, pharmacokinetic or pharmacodynamic properties.
Isoniazid (INH), a first-line anti-TB drug, is one of the most effective agents that has been used for the treatment of Mycobacterium tuberculosis infection since 1952.25,26 It is a pro-drug which is activated by the mycobacterial catalase–peroxidase enzyme known as KatG. The activated form reacts with the coenzyme NADH to form an isonicotinic acyl–NADH complex27,28 that binds with the enoyl–acyl carrier protein (ACP) reductase InhA, which is involved in the elongation of fatty acids during the mycolic acid synthesis.29 Thus isoniazid inhibits the synthesis of mycolic acid, required for the mycobacterial cell wall. INH is metabolized in the liver and forms compounds such as hydrazine, which are toxic to the central nervous system and other organs.30,31 Because INH is an important drug in the therapeutic arsenal for TB treatment, efforts are being made to develop new INH derivatives with greater activity, lower toxicity and fewer side effects than INH.32–41 Several recent reports indicate that the incorporation of hydrophobic moieties into the basic structure of INH can enhance penetration of the drug into the highly lipophilic cell wall of the bacterium. Moreover, by functionalizing the hydrazine group of isoniazid and retaining its activity we can avoid the toxicity and other severe problems related to the inactivation of isoniazid by the enzyme N-acetyltransferase-2 (NAT2).
Several research groups have introduced the amido ether functionality into biologically active molecules and the resulting hybrids exhibited good biological activities.42–46 Encouraged by the previous studies and in continuation of our efforts towards the synthesis of new anti-tuberculosis agents,47–51 we proposed to attach an amido ether linkage to isoniazid to form isoniazid–amidoether derivatives and evaluated their in vitro and in vivo anti-TB activity and toxicity against THP-1 cell lines.
Chemistry
For the synthesis of isoniazid–amidoether conjugates (3a–3v), first different 2-(4-formylphenoxy)-N-substituted acetamides (2a–2v) were synthesized as shown in Scheme 1. The synthesis started with the reaction of 2-chloroacetyl chloride and an appropriate amine in the presence of triethylamine as a base in dichloromethane, that leads to the formation of chloroacetamide derivatives (1a–1v) in quantitative yields (Scheme 1).52 These derivatives were then treated with p-hydroxy benzaldehyde in the presence of K2CO3 as the base and potassium iodide as the catalyst to give 2-(4-formylphenoxy)-N-substituted acetamides (2a–2v). These compounds with a free carboxyl group were then condensed with isoniazid in ethanol–H2O as a solvent to get the desired compounds (3a–3v) in good yields. All the synthesized compounds were purified by column chromatography and characterized spectroscopically.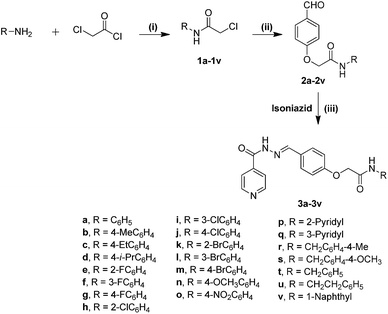 | ||
| Scheme 1 Reagents and conditions: (i) Et3N, DCM, 0 °C to RT, 6–10 h, 70–90%; (ii) 4-hydroxybenzaldehyde, K2CO3, KI, RT, 10–15 h, 60–85%; (iii) EtOH–H2O, RT, 6–10 h, 70–90%. | ||
Biological activity
In vitro anti-tuberculosis activity
A stock culture of M. tb H37Rv (ATCC 27294) was grown to an Abs600 nm of 0.2 in Middlebrook 7H9 broth (Difco) supplemented with 0.05% Tween 80, 0.2% glycerol and an albumin–NaCl–glucose (ADS) complex. The culture was diluted 1 : 1000 in 7H9-based medium before aliquoting 50 μL into each well of a 96-well plate. Compounds were dissolved in DMSO (Sigma) to make stock solutions of 50 mM. Compounds (100 μL solution) were added to the first row of the 96-well plate at a final concentration of 100 μM. 2-fold serial dilutions were made and 5 dilutions of each compound (50 μM–0.195 μM) were tested for anti-mycobacterial activity. The compounds were diluted 1 : 1 by the addition of 50 μL of 1 : 1000 diluted cultures. Rows 6 and 12 of the 96-well plates were controls with no compound. The plates were incubated at 37 °C and the MIC99 values were read macroscopically using an inverted plate reader after 14 days. MIC99 is defined as the minimum inhibitory concentration of the compound required for 99% inhibition of bacterial growth. Each measurement was repeated thrice.In vivo anti-tuberculosis activity
The most active compound 3b from the series was selected for in vivo antituberculosis activity evaluation. For activity evaluation, pathogen-free Balb/c mice of either sex (25–30 g) were procured from the Division of Laboratory Animals, Central Drug Research Institute, Lucknow, India. The animals were maintained in a BSLIII animal facility at the University of Delhi South Campus, New Delhi and routinely cared according to the guidelines of CPCSEA (Committee for the Purpose of Control and Supervision of Experiments on Animals), India. All the experimental protocols included in this study were reviewed and approved by the Institutional Animal Ethics Committee (Ref no. 1/IAEC/AKT/Biochem/UDSC/14.10.2011).Mice were tested with M. tb. H37Rv bacilli by the respiratory route in an inhalation chamber (Glascol Inc., USA) pre-calibrated to deliver approximately 1000 bacilli per animal in the lung by using frozen stocks of M. tb. H37Rv with their CFU pre-determined. Mice were euthanized on the day after infection to determine the number of CFUs implanted in the lungs. Following two weeks of infection, mice were divided into different groups – untreated, DMSO, isoniazid (25 mg kg−1), rifampicin (10 mg kg−1), and compound 3b (25 mg kg−1, 50 mg kg−1, 100 mg kg−1) and therapy was initiated.
All drugs were administered once daily, five days per week, in a maximum volume of 0.175 mL by oral gavage. Before initiating the chemotherapy, the infection in the animals was verified by euthanizing a group of animals (N = 5) at 2 weeks post-infection followed by pathological observation and an enumeration of the bacillary load in the lungs and spleens. Mice (N = 5 each group) were euthanized by CO2 asphyxiation after three week, six week and ten week time points post-therapy and monitored for gross pathological observations and bacillary load. For bacterial enumeration, mice were dissected and lungs and spleens were aseptically removed and homogenized in saline. Appropriate dilutions of the homogenates were plated in duplicate onto MB7H11 agar and the plates were incubated at 37 °C for 3–4 weeks followed by an enumeration of colonies. The results were expressed as log10 CFU per organ.
At the 3 week time point, a bacillary load of 6.38 and 4.19 log10 CFU was measured in the lungs and spleens of untreated animals. The disease continued to persist and at 6 weeks post-infection also, a bacillary load of 5.91 and 3.68 log10 was recorded in the lungs and spleens of untreated animals, respectively. At 10 weeks post-infection, the disease further progressed with a bacillary load of 6.04 and 4.02 log10 in the lungs and spleens of untreated animals, respectively.
Results and discussion
The isoniazid amidoether derivatives (3a–3v) synthesized as shown in Scheme 1 were evaluated for antitubercular activity against Mycobacterium tuberculosis H37Rv using isoniazid as the reference compound. The minimum inhibitory concentrations (MIC99) of the compounds are shown in Table 1. Five compounds (3b, 3n, 3q, 3r and 3s) showed excellent activity with MIC99 = 0.39 μM similar to isoniazid. In general, compounds having electron donating groups at the para position of the benzene ring such as 3n (C6H4-4-OMe), 3r (CH2C6H4-4-Me), 3s (CH2C6H4-4-OMe) and 3b–3d (alkyl groups) were found to be more active in comparison to other compounds. Electron withdrawing groups lead to a slight decrease in activity, as compounds 3o (4-NO2) and 3v (1-naphthyl) showed weak activity (MIC99 = 1.56–3.125 μM) compared to other compounds.| Comp. | MIC99 (μM) | THP-1 (μM) | Clog P | |
|---|---|---|---|---|
| R | ||||
| 3a | C6H5 | 0.78 | >50 | 2.30 |
| 3b | 4-MeC6H4 | 0.39 | >50 | 2.80 |
| 3c | 4-EtC6H4 | 0.39–0.78 | >50 | 3.33 |
| 3d | 4-i-PrC6H4 | 0.39–0.78 | >50 | 3.73 |
| 3e | 2-FC6H4 | 0.78 | >50 | 2.10 |
| 3f | 3-FC6H4 | 0.78 | >50 | 2.70 |
| 3g | 4-FC6H4 | 0.78 | >50 | 2.70 |
| 3h | 2-ClC6H4 | 0.78 | >50 | 2.42 |
| 3i | 3-ClC6H4 | 0.78 | >50 | 3.27 |
| 3j | 4-ClC6H4 | 0.39–0.78 | >50 | 3.27 |
| 3k | 2-BrC6H4 | 0.78 | >50 | 2.54 |
| 3l | 3-BrC6H4 | 0.78 | >50 | 3.42 |
| 3m | 4-BrC6H4 | 0.78 | >50 | 3.42 |
| 3n | 4-OCH3C6H4 | 0.39 | >50 | 2.38 |
| 3o | 4-NO2C6H4 | 1.56–3.125 | >50 | 2.60 |
| 3p | 2-Pyridyl | 0.78 | >50 | 1.63 |
| 3q | 3-Pyridyl | 0.39 | >50 | 1.63 |
| 3r | CH2C6H4-4-Me | 0.39 | >50 | 2.83 |
| 3s | CH2C6H4-4-OCH3 | 0.39 | >50 | 2.25 |
| 3t | CH2C6H5 | 0.78 | >50 | 2.33 |
| 3u | CH2CH2C6H5 | 0.78 | >50 | 2.54 |
| 3v | 1-Naphthyl | 1.56–3.125 | >50 | 3.48 |
| Isoniazid | 0.39 | −0.668 | ||
After 3 weeks of treatment with isoniazid, the CFU in the lungs of the infected animals was reduced by 1.22 log10 while after 6 weeks it showed a further CFU reduction in the bacillary load by 2.59 log10 and a further reduction of 3.38 log10 at 10 weeks when compared with the animals in the untreated group (Fig. 1A). In spleens also, isoniazid treatment resulted in a sharp reduction of bacillary load with a 2.66 log10 reduction at 3 weeks when compared with the animals in the untreated group (Fig. 1B). At 6 week and 10 week time points, no bacilli were recovered from the spleens. After 3 weeks of treatment with rifampicin, the CFU in the lungs of the infected animals was reduced by 1.93 log10 while at 6 weeks they showed a further CFU reduction in the bacillary load by 2.8 log10 when compared with the animals in the DMSO group. After 10 weeks of rifampicin administration, no bacilli were recovered from the lungs of the infected animals (Fig. 1A). In spleens also, rifampicin treatment resulted in a marked reduction in the bacillary load with a 1.76 log10 and 3.31 log10 reduction at 3 weeks and 6 weeks, respectively when compared with the animals in the DMSO group. At the 10 week time point, no bacilli were recovered from the spleens (Fig. 1B).
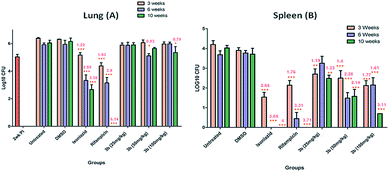 | ||
| Fig. 1 Bacterial enumeration in lungs (A) and spleens (B) of animals belonging to different groups was carried out at various time points following infection. The bacillary load was measured as described in the Materials and methods section. Various groups are indicated. PI represents post-infection. ***, ** and * indicate statistical significance with P values <0.001, <0.01 and <0.05, respectively. The numbers in red represent the log10 CFU value by which a reduction in the particular case was observed when compared with the CFU value in the case of control animals. | ||
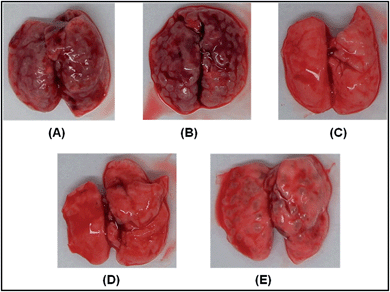
Compound 3b was administered at 25 mg kg−1, 50 mg kg−1 and 100 mg kg−1 concentrations for evaluating the efficacy of this compound. When compound 3b was given at a low concentration of 25 mg kg−1, it exhibited no control towards disease progression even up to 10 weeks as was evident from a comparable bacillary load observed in the case of lungs of animals treated with compound 3b as compared to DMSO treated animals. When the concentration of compound 3b was increased to 50 mg kg−1 or 100 mg kg−1 still no significant influence of the compound towards pulmonary control of the disease was observed hence, in spite of increasing the concentration up to 100 mg kg−1, no significant chemotherapeutic effect was demonstrable under our experimental conditions. At 50 mg kg−1 concentration of the compound at the 6 week time point and at 100 mg kg−1 concentration at the 10 week time point, there was a slight reduction in the pulmonary bacillary load of compound 3b treated animals as compared to DMSO treated animals, however, the statistical significance was not of a very high order and the fact that it did not register any significant chemotherapeutic effect even at 100 mg kg−1 concentration shows that the compound had no significant intrinsic chemotherapeutic value towards the control of pulmonary tuberculosis at least in the murine model as observed in our experiments. Fig. 2 depicts the lungs of infected mice either untreated or treated with isoniazid, rifampicin and compound 3b (100 mg kg−1) for 10 weeks.
After the pulmonary infection, the settlement of bacilli in the lung tissues is followed by hematogenous spread which provides another point of control towards protection from the disease hence, the influence of compound 3b was also measured on the reduction of bacillary load in spleens.
It was observed that at 25 mg kg−1 concentration of compound 3b, there was a reduction in the splenic bacillary load by a value of 1.19 log10 CFU at the end of 3 weeks of chemotherapy when compared with the DMSO treated animals, however, treatment up to 10 weeks further reduced the splenic bacillary load only marginally with 1.23 log10 CFU reduction in the bacillary load at this time point (Fig. 1B). With increased concentration of compound 3b to 50 mg kg−1, a more substantial effect was observed on the reduction of splenic bacillary load. We observed that at this concentration, at the end of 3 weeks of chemotherapy the splenic bacillary load exhibited a reduction by 1.4 log10 CFU. Further continuation with the chemotherapy at this dose i.e. up to 6 weeks and 10 weeks resulted in more significant reduction in the splenic bacillary load which was 2.28 log10 CFU and 2.14 log10 CFU less compared to the bacillary load in the spleens of DMSO treated animals. When the concentration of compound 3b was doubled up to a concentration of 100 mg kg−1, it resulted in a more prominent bacillary load reduction in spleens and as compared to 1.4 log10 CFU reduction observed with 50 mg kg−1 concentration, this concentration resulted in a 1.77 log10 CFU reduction in the splenic bacillary load at the end of 3 weeks of treatment. Further administration of the compound to 10 weeks resulted in a very significant 3.11 log10 CFU reduction in the splenic bacillary load compared to the bacillary load in the spleens of DMSO treated animals (Fig. 1B). All the compounds were further examined for toxicity in the THP-1 cell line. The compounds were found to be non-toxic up to a concentration of 50 μM, the highest concentration tested (Table 1). It was not possible to test toxicity at higher concentrations due to solubility limitations.
In silico ADMET prediction
We have predicted the ADME properties of test compounds and reference compound isoniazid for the pharmaceutically relevant properties to assess the drug likeness and pharmacokinetic properties. The Qikprop v3.5 (Schrödinger, Inc., New York, NY, 2012) was used for the evaluation of some important absorption, distribution, metabolism and elimination (ADME) parameters and its permissible ranges are listed in Tables 2 and 3.| Comp. | Mol_MW (>500) | Donor HB (<5) | Accpt. HB (<10) | QPlog Po/w (<5) | Rule of Five (<4) |
|---|---|---|---|---|---|
| a All values calculated by QikProp v 3.5 and the explanations of the descriptors are given in the text. | |||||
| 3a | 374.39 | 2 | 7.25 | 3.139 | 0 |
| 3b | 388.42 | 2 | 7.25 | 3.347 | 0 |
| 3c | 402.45 | 2 | 7.25 | 3.621 | 0 |
| 3d | 416.47 | 2 | 7.25 | 4.272 | 0 |
| 3e | 392.38 | 2 | 7.25 | 3.329 | 0 |
| 3f | 392.38 | 2 | 7.25 | 3.343 | 0 |
| 3g | 392.38 | 2 | 7.25 | 3.577 | 0 |
| 3h | 408.84 | 2 | 7.25 | 3.576 | 0 |
| 3i | 408.84 | 2 | 7.25 | 3.619 | 0 |
| 3j | 408.84 | 2 | 7.25 | 3.904 | 0 |
| 3k | 453.29 | 2 | 7.25 | 3.89 | 0 |
| 3l | 453.29 | 2 | 7.25 | 3.93 | 0 |
| 3m | 453.29 | 2 | 7.25 | 4.078 | 0 |
| 3n | 404.42 | 2 | 8 | 3.362 | 0 |
| 3o | 419.39 | 2 | 8.25 | 2.814 | 0 |
| 3p | 375.38 | 2 | 8.25 | 2.832 | 0 |
| 3q | 375.38 | 2 | 8.75 | 2.59 | 0 |
| 3r | 402.45 | 2 | 7.25 | 3.206 | 0 |
| 3s | 418.45 | 2 | 8 | 3.334 | 0 |
| 3t | 388.42 | 2 | 7.25 | 3.106 | 0 |
| 3u | 402.45 | 2 | 7.25 | 3.593 | 0 |
| 3v | 424.45 | 2 | 7.25 | 4.46 | 0 |
| INH | 137.14 | 3 | 4.5 | −0.646 | 0 |
| Compd | Percent human oral absorptiona (>80% high, <25% poor) | QPPCacoa nm s−1 (<25 poor, >500 great) | QPlog BBa (−3.0 to 1.2) | QPPMDCKa (<25 poor, >500 great) | QPlog Khsaa (−1.5 to 1.5) | PSAa (7.0–200.0) | #Rotora (0–15) |
|---|---|---|---|---|---|---|---|
| a Calculated using QikProp v 3.5. Range/recommended values calculated for 95% known drugs. | |||||||
| 3a | 91.362 | 373.559 | −1.66 | 170.656 | 0.195 | 107.913 | 8 |
| 3b | 92.58 | 373.506 | −1.702 | 170.63 | 0.345 | 107.914 | 8 |
| 3c | 94.137 | 371.023 | −1.799 | 169.404 | 0.451 | 107.915 | 9 |
| 3d | 100 | 370.991 | −1.819 | 169.388 | 0.584 | 107.917 | 9 |
| 3e | 93.157 | 407.696 | −1.523 | 282.274 | 0.225 | 107.448 | 8 |
| 3f | 92.52 | 371.547 | −1.559 | 306.467 | 0.236 | 107.923 | 8 |
| 3g | 93.934 | 373.725 | −1.557 | 308.982 | 0.235 | 107.912 | 8 |
| 3h | 95.15 | 437.44 | −1.428 | 411.765 | 0.291 | 106.704 | 8 |
| 3i | 94.178 | 373.705 | −1.516 | 420.73 | 0.304 | 107.912 | 8 |
| 3j | 95.849 | 373.693 | −1.515 | 421.519 | 0.304 | 107.913 | 8 |
| 3k | 100 | 454.177 | −1.408 | 463.239 | 0.308 | 107.1 | 8 |
| 3l | 95.997 | 373.715 | −1.509 | 452.415 | 0.326 | 107.912 | 8 |
| 3m | 96.867 | 373.649 | −1.509 | 453.167 | 0.327 | 107.913 | 8 |
| 3n | 92.665 | 373.326 | −1.757 | 170.541 | 0.201 | 116.206 | 9 |
| 3o | 72.954 | 44.669 | −2.925 | 17.186 | 0.146 | 152.833 | 9 |
| 3p | 86.926 | 265.849 | −1.82 | 118.154 | −0.01 | 119.081 | 8 |
| 3q | 83.384 | 202.307 | −1.976 | 87.949 | −0.116 | 120.825 | 8 |
| 3r | 86.5 | 189.976 | −1.803 | 165.521 | 0.135 | 109.005 | 9 |
| 3s | 87.243 | 189.824 | −1.858 | 165.336 | −0.011 | 117.291 | 10 |
| 3t | 85.908 | 189.805 | −1.763 | 165.453 | −0.016 | 108.999 | 9 |
| 3u | 89.562 | 210.386 | −1.86 | 165.226 | 0.088 | 109.683 | 10 |
| 3v | 100 | 451.89 | −1.601 | 209.643 | 0.528 | 105.996 | 8 |
| Isoniazid | 66.893 | 277.461 | −0.843 | 123.742 | −0.752 | 81.355 | 2 |
All the compounds were prepared in neutralized form for the calculation of pharmacokinetic properties by Maestro Build module and LigPrep, saved in SD format. In the present study, the test compounds showed good drug-like properties based on Lipinski's rule of 5 showing zero violation of the rule, proving all the test compounds to be orally active. The descriptor QPPCaco indicating Caco-2 cells permeability, a model used for the gut–blood barrier, showed good values for all the test compounds. Similarly the values of the descriptor model such as the number of rotatable bonds (#rotor) and polar surface area (PSA), used as an indicator of bioavailability for the test compounds, lie in the expected ranges. Further, the prediction for human serum albumin binding (QPlog Khsa) and the QikProp descriptor for the brain–blood partition coefficient (QPlog BB) and the blood–brain barrier mimic MDCK cell permeability (QPPMDCK) show satisfactory predictions for all the test compounds (Table 3).
Conclusion
We have synthesized 22 isoniazid–amidoether conjugates and evaluated their in vitro and in vivo anti-tuberculosis activity. Most of the compounds exhibited potent activity in vitro. When tested for in vivo activity, the compound exhibited mild activity in the case of lungs. However, the influence of the compound on the replication of the pathogen in the spleen was far superior as compared to the influence on the lungs. We do not have clear explanation for this observation at present however it might suggest that the bio-availability of these compounds could be the possible reason for the better activity profile in the case of spleens. However, translation of this speculation into real evidence would require further experiments. All the compounds were found to be nontoxic up to 50 μM concentration against the THP-1 cell line. Also, the compounds exhibited good pharmacokinetic properties and follow Lipinski's rule of 5. Thus we believe that these compounds can be considered as a possible lead for the development of new anti-tuberculosis agents.Acknowledgements
D.S.R. thanks the Council of Scientific and Industrial Research [no. 02(0049)/12/EMR-II] New Delhi, India for financial support. A.K.T. is thankful to the Department of Biotechnology [no. BT/01/COE/05/06-II] Ministry of Science and Technology, Government of India for financial support. D.K. and Beena are thankful to CSIR for the award of junior and senior research fellowship. Authors are thankful to Prija Ponnan for the in silico ADMET prediction studies. Authors are also thankful to CIF–USIC, University of Delhi, Delhi for NMR spectral data and RSIC, CDRI, Lucknow for mass data.Notes and references
- http://whqlibdoc.who.int/publications/2011/9789241564380_eng.pdf .
- Beena and D. S. Rawat, Med. Res. Rev., 2013, 33, 693 CrossRef PubMed.
- G. B. Migliori, G. De Iaco, G. Besozzi, R. Centis and D. M. Cirillo, Euro. Surveill., 2007, 12, E070517.1 Search PubMed.
- A. A. Velayati, M. R. Masjedi, P. Farnia, P. Tabarsi, J. Ghanavi and A. H. ZiaZarifi, Chest, 2009, 136, 420 CrossRef PubMed.
- K. Rowland, Nature, 2012, http://www.nature.com/news/totally-drugresistant- tb-emerges-in-india-1.9797 Search PubMed.
- S. Loewenberg, Lancet, 2012, 379, 205 CrossRef.
- A. S. Fauci, J. Infect. Dis., 2008, 197, 1493 CrossRef PubMed.
- S. Swaminathan, C. Padmapriyadarsini and G. Narendran, Clin. Infect. Dis., 2010, 50, 1377 CrossRef CAS PubMed.
- R. A. M. Breen, L. Swaden, J. Ballinger and M. C. I. Lipman, Drugs, 2006, 66, 2299 CrossRef CAS PubMed.
- S. K. Jain, G. Lamichhane, S. Nimmagadda, M. G. Pomper and W. R. Bishai, Microbe, 2008, 3, 285 Search PubMed.
- P. Ahirrao, Mini-Rev. Med. Chem., 2008, 8, 1441 CrossRef CAS PubMed.
- B. Villemagne, C. Crauste, M. Flipo, A. R. Baulard, B. Deprez and N. Willand, Eur. J. Med. Chem., 2012, 51, 1 CrossRef CAS PubMed.
- A. M. Ginsberg, Tuberculosis, 2010, 90, 162 CrossRef CAS PubMed.
- S. T. Cole and G. Riccardi, Curr. Opin. Microbiol., 2011, 14, 570 CrossRef CAS PubMed.
- P. Ahirrao, Mini-Rev. Med. Chem., 2008, 8, 1441 CrossRef CAS PubMed.
- http://www.newtbdrugs.org/pipeline.php .
- A. M. Ginsberg, Tuberculosis, 2010, 90, 162 CrossRef CAS PubMed.
- S. T. Cole and G. Riccardi, Curr. Opin. Microbiol., 2011, 14, 570 CrossRef CAS PubMed.
- A. H. Diacon, P. R. Donald, A. Pym, M. Grobusch, R. F. Patientia, R. Mahanyele, N. Bantubani, R. Narasimooloo, T. De Marez, R. van Heeswijk, N. Lounis, P. Meyvisch, K. Andries and D. F. McNeeley, Antimicrob. Agents Chemother., 2012, 56, 3271 CrossRef CAS PubMed.
- R. Mahajan, Int. J. Appl. Basic Med. Res., 2013, 3, 1 CrossRef PubMed.
- M. S. Butler, M. A. Blaskovich and M. A. Cooper, J. Antibiot., 2013, 66, 571 CrossRef CAS PubMed.
- D. J. Knowles, Trends Microbiol., 1997, 5, 379 CrossRef CAS PubMed.
- C. R. Harris and A. Thorarensen, Curr. Med. Chem., 2004, 11, 2213 CrossRef CAS PubMed.
- I. Chopra, J. Hodgson, B. Metcalf and G. Poste, JAMA, 1996, 275, 401 CrossRef CAS PubMed.
- H. A. Offe, W. Siefken and G. Domagk, Naturwissenschaften, 1952, 39, 118 CrossRef CAS.
- E. Grunberg and R. J. Schnitzer, Proc. Soc. Exp. Biol. Med., 1953, 84, 220 CrossRef CAS PubMed.
- X. Zhao, H. Yu, S. Yu, F. Wang, J. C. Sacchettini and R. S. Magliozzo, Biochemistry, 2006, 45, 4131 CrossRef CAS PubMed.
- M. Nguyen, C. Claparols, J. Bernadou and B. Meunier, ChemBioChem, 2001, 2, 877 CrossRef CAS PubMed.
- R. Rawat, A. Whitty and P. J. Tonge, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 13881 CrossRef CAS PubMed.
- T. C. Sarich, M. Youssefi, T. Zhou, S. P. Adams, R. A. Wall and J. M. Wright, Arch. Toxicol., 1996, 70, 835 CrossRef CAS PubMed.
- G. A. Ellard and P. T. Gammon, J. Pharmacokinet. Biopharm., 1976, 4, 83 CrossRef CAS PubMed.
- R. Maccari, R. Ottana, F. Monforte and M. G. Vigorita, Antimicrob. Agents Chemother., 2002, 46, 294 CrossRef CAS PubMed.
- T. H. Manjashetty, P. Yogeeswari and D. Sriram, Bioorg. Med. Chem. Lett., 2011, 21, 2125 CrossRef CAS PubMed.
- A. V. Ramani, A. Monika, V. L. Indira, G. Karyavardhi, J. Venkatesh, V. U. Jeankumar, T. H. Manjashetty, P. Yogeeswari and D. Sriram, Bioorg. Med. Chem. Lett., 2012, 22, 2764 CrossRef CAS PubMed.
- E. Vavrikova, S. Polanc, M. Kocevar, K. Horvati, S. Bosze, J. Stolarikova, K. Vavrova and J. Vinsova, Eur. J. Med. Chem., 2011, 46, 4937 CrossRef CAS PubMed.
- E. Vavrikova, S. Polanc, M. Kocevar, J. Kosmrlj, K. Horvati, S. Bosze, J. Stolarikova, A. Imramovsky and J. Vinsova, Eur. J. Med. Chem., 2011, 46, 5902 CrossRef CAS PubMed.
- N. Boechat, V. F. Ferreira, S. B. Ferreira, M. L. G. Ferreira, F. C. Silva, M. M. Bastos, M. S. Costa, M. C. S. Lourenco, A. C. Pinto, A. U. Krettli, A. C. Aguiar, B. M. Teixeira, N. V. Silva, P. R. C. Martins, F. A. F. M. Bezerra, A. L. S. Camilo, G. P. Silva and C. C. P. Costa, J. Med. Chem., 2011, 54, 5988 CrossRef CAS PubMed.
- P. De, G. K. Yoya, P. Constant, F. B. Belval, H. Duran, N. Saffon, M. Daffe and M. Baltas, J. Med. Chem., 2011, 54, 1449 CrossRef CAS PubMed.
- K. Elumalai, M. A. Ali, M. Elumalai, K. Eluri and S. Srinivasan, J. Acute Disease, 2013, 316 CrossRef.
- L. Matei, C. Bleotu, I. Baciu, C. Draghici, P. Ionita, A. Paun, M. C. Chifiriuc, A. Sbarcea and I. Zarafu, Bioorg. Med. Chem., 2013, 21, 5355 CrossRef CAS PubMed.
- F. Martins, S. Santos, C. Ventura, R. Elvas-Leitao, L. Santos, S. Vitorino, M. Reis, V. Miranda, H. F. Correia, J. Aires-de-Sousa, V. Kovalishyn, D. A. R. S. Latino, J. Ramos and M. Viveiros, Eur. J. Med. Chem., 2014, 81, 119 CrossRef CAS PubMed.
- G. Jin, D. Lu, S. Yao, C. C. Wu, J. X. Liu, D. A. Carson and H. B. Cottam, Bioorg. Med. Chem. Lett., 2009, 19, 606 CrossRef CAS PubMed.
- S. S. El-Nakkady, M. M. Hanna, H. M. Roaiah and I. A. Y. Ghannam, Eur. J. Med. Chem., 2012, 47, 387–398 CrossRef CAS PubMed.
- S. Fortin, E. Moreau, J. Lacroix, M. F. Cote, E. Petitclerc and R. C. Gaudreault, Eur. J. Med. Chem., 2010, 45, 2928 CrossRef CAS PubMed.
- L. Maa, S. Li, H. Zheng, J. Chen, L. Lin, X. Ye, Z. Chen, Q. Xu, T. Chen, J. Yang, N. Qiu, G. Wang, A. Peng, Y. Ding, Y. Wei and L. Chen, Eur. J. Med. Chem., 2001, 46, 2003 CrossRef PubMed.
- L. Ma, J. Chen, X. Liang, C. Xie, C. Deng, L. Huang, A. Peng, Y. Wei and L. Chen, Arch. Pharm. Chem. Life Sci., 2012, 345, 517 CrossRef CAS PubMed.
- Beena, S. Joshi, N. Kumar, S. Kidwai, R. Singh and D. S. Rawat, Arch. Pharm. Chem. Life Sci., 2012, 345, 896 CrossRef CAS PubMed.
- D. Kumar, K. K. Raj, M. Bailey, T. Alling, T. Parish and D. S. Rawat, Bioorg. Med. Chem. Lett., 2013, 23, 1365 CrossRef CAS PubMed.
- Beena, D. Kumar, W. Kumbukgolla, S. Jayaweera, M. Bailey, T. Alling, J. Ollinger, T. Parish and D. S. Rawat, RSC Adv., 2014, 4, 11962 RSC.
- Beena, D. Kumar, M. Bailey, T. Parish and D. S. Rawat, Chem. Biol. Interface, 2014, 4, 23 Search PubMed.
- D. Kumar, Beena, G. Khare, S. Kidwai, A. K. Tyagi, R. Singh and D. S. Rawat, Eur. J. Med. Chem., 2014, 81, 301 CrossRef CAS PubMed.
- P. G. Baraldi, D. Preti, M. A. Tabrizi, F. Fruttarolo, G. Saponaro, S. Baraldi, R. Romagnoli, A. R. Moorman, S. Gessi, K. Varanib and P. A. Borea, Bioorg. Med. Chem., 2007, 15, 2514 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4md00288a |
| This journal is © The Royal Society of Chemistry 2015 |