DOI:
10.1039/C4MD00266K
(Concise Article)
Med. Chem. Commun., 2015,
6, 147-154
Synthesis of tetrazole compounds as a novel type of potential antimicrobial agents and their synergistic effects with clinical drugs and interactions with calf thymus DNA†
Received 21st June 2014 , Accepted 22nd September 2014
First published on 22nd September 2014
1. Introduction
Microbial infections have become alarming recently due to the increasing emergence of multidrug-resistant strains, intractable pathogenic microorganisms and newly arising pathogens.1–4 Various synthetic and semi-synthetic antimicrobial agents have been discovered and extensively used in the clinic to treat various community and hospital acquired microbial infections. Especially, the FDA approved drug COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSauranofin exerts an incredible effect on Gram-positive and multidrug-resistant bacteria and has attracted much attention recently.5,6 However, there are still some unresolved problems such as narrow antimicrobial spectrum, adverse effects, high toxicity and so on for clinical drugs. The combination therapy with two or more agents, which could usually overcome multi-drug resistance, is one of the important strategies to improve the efficiency and bioavailability as well as treat mixed diseases in the clinic.7,8 However, the discovery and development of structurally novel antimicrobial agents with good pharmacological profiles and excellent activity towards resistant strains are highly desirable.9–11 COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSTetrazole is an important five-membered aromatic heterocyclic compound with poly-nitrogen electron-rich planar structural features. This unique structure allows tetrazole derivatives to readily bind with various enzymes or receptors in organisms via weak interactions such as coordination bonds, hydrogen bonds, cation–π, π–π stacking, hydrophobic effect, van der Waals force and so on, thus displaying a broad spectrum of biological activities and a considerable role in the pharmaceutical field.12,13 The tetrazole ring can be used as an attractive linker to combine or stabilize different pharmacophore fragments to generate special functionalized molecules.14,15 The tetrazole ring is also an isostere of carboxyl,16,17 amide18 and some heterocycles (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTStriazole,19 benzotriazole,20 COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSimidazole,21 COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSbenzimidazole,22 COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTScarbazole,23etc.) in designing various new types of drug molecules. So far many COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTStetrazole-based derivatives have been successfully developed and prevalently used as clinical drugs such as antihypertensives Lorsartan24 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSValsartan,25 antibiotics Flomoxef26 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSCefonicid,27 and antinociceptive COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSAlfentanil28 to treat various diseases. Some literature has revealed that tetrazole derivatives could effectively prevent the biosynthesis of microbial proteins29,30 to inhibit the growth of various microorganisms in recent years, which suggests the large potentiality of tetrazole compounds as a new type of antimicrobial agents.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole is one of the most important COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTStriazole-based compounds recommended as the first-line antifungal drug by the World Health Organization (WHO). It has been prevalently employed to treat fungal infection by Candida albicans, Cryptococcus neoformans, Dermatitis blastomycosis, etc. due to its potent activity, excellent safety profile, and favorable pharmacokinetic characteristics.31,32 The triazole ring of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole could efficiently coordinate with the iron(II) ion of heme to inhibit the biosynthesis of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSergosterol and thus inhibits the growth of fungi. However, the emergence of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSfluconazole-resistant Candida albicans isolates, increasingly serious drug resistance, narrow antifungal spectrum, and low activity against invasive mycoses have attracted great efforts towards modifying the side chain of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole or exploiting its new analogues.33 In our previous work,34–36 it has been demonstrated that the tertiary amine type of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole analogues displayed large potentiality as a new type of antimicrobial agents in which the tertiary alcohol moiety in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole was replaced by a tertiary amine fragment that may exert the same function with the active site residue H310 as the tertiary alcohol moiety in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole. However, to the best of our knowledge, so far the combination of a tetrazole ring with a tertiary amine fragment has not been reported.
In view of the above considerations and as an extension of our continuous work, we have great interest in investigating COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTStetrazole tertiary amines as a novel type of potential antimicrobial agents. Herein, we would like to report the synthesis of tetrazole compounds 4a–d and 6a–l with different lengths of alkyl chains or halogen-substituted aralkyl moieties. All the new compounds were screened for their antibacterial and antifungal activities in vitro, and the synergistic effects of the most active tetrazole compound with clinical drugs were also evaluated in vitro. The ionization constants (pKa) and lg P of title compounds were determined by the UV-vis absorption spectroscopic method to evaluate the antimicrobial activity. In this work, with the consideration to explore the preliminary mechanism of action, the interaction of the most active compound with calf thymus DNA was also investigated.
2. Chemistry
The target tetrazole compounds 4a–d and 6a–l were prepared via multi-step reactions and the synthetic process was outlined in Scheme 1. Commercially available substituted anilines were treated by COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSchloroacetonitrile in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSacetonitrile to give N-phenyl glycinonitriles 2a–c in 50–65% yields. The N-alkylation of compounds 2a–c with a series of alkyl bromides in refluxing COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSethanol using potassium carbonate as the base respectively afforded the intermediates 3a–d in yields of 50–55%, which showed that the length of the aliphatic chain in alkyl bromides exerted a slight effect on the formation of tertiary amines. Halobenzyl halides and intermediates 2a–c underwent N-alkylation to produce compounds 5a–l in yields of 65–70%. Compounds 4a–d and 6a–l were easily synthesized in high yields ranging from 75% to 80% by the cycloaddition of compounds 3a–d and 5a–l respectively with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSsodium azide using COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSammonium chloride as the catalyst in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSDMF at 120 °C. The total yields of target compounds were ranging from 18% to 36%. Moreover, in the procedure for the preparation of N-phenyl glycinonitriles, it was found that no anticipated products were observed when the aniline ring was substituted by electron-withdrawing groups such as fluoro, chloro, nitro and so on. This phenomenon might be attributed to the electron-withdrawing properties of these groups which made the electron density of the nitrogen atom in the aniline ring low and made it difficult to perform the nucleophilic substitution. All the new compounds were confirmed by IR,1H NMR, 13C NMR, MS and HRMS spectroscopy (See the ESI 1†). 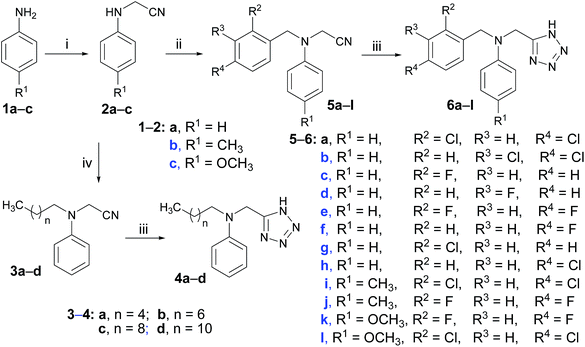 |
| | Scheme 1 Reagents and conditions: (i) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSchloroacetonitrile, potassium carbonate, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSCH3CN, 80 °C; (ii) substituted halobenzyl halide, potassium carbonate, CH3CH2OH, reflux; (iii) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSsodium azide, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSammonium chloride, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSDMF, 120 °C; (iv) alkyl bromide, potassium carbonate, CH3CH2OH, reflux. | |
3. Results and discussion
The results of antibacterial activity (ESI 2†) in Table 1 showed that intermediates 3a–d and 5a–l exhibited weak or no antibacterial efficacy against all the tested bacterial strains. In addition, some of the halobenzyl tertiary amines among 5a–l exerted relatively better activities in inhibiting the growth of tested strains in comparison with alkyl tertiary amines 3a–d. Moreover, the bioactivity of target compounds 4a–d and 6a–l were improved by the introduction of a tetrazole ring into substituted benzyl tertiary amines, which indicated that the tetrazole ring was helpful for enhancing antibacterial activity. The types of substituents on the aniline ring exhibited no significant effects on biological activity. Moreover, substituents on benzyl moieties displayed notable effects on antibacterial activity, especially COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g (MIC = 16–32 μg mL−1) with the chlorine atom on the 2-position of the benzyl ring gave better bioactivity against all the tested bacterial strains than others.
Table 1 Antimicrobial activities in vitro for the prepared compounds expressed as MIC (μg mL−1)a,b,c
| Compds | Gram-positive bacteria | Gram-negative bacteria | Fungi |
| S. A | MRSA | B. S | M. L | B. P | E. C | P. A | S. D | C. A | C. M | C. U | A. F |
Minimum inhibitory concentrations were determined by micro broth dilution method for microdilution plates. MRSA, Methicillinresistant Staphylococcus aureus N315; S. A, Staphylococcus aureus ATCC25923; B. S, Bacillus subtilis ATCC6633; M. L, Micrococcus luteus ATCC 4698; E. C, Escherichia coli DH52; S. D, Shigella dysenteriae ATCC51252; P. A, Pseudomonas aeruginosa ATCC27853; B. P, Bacillus proteus ATCC13315; C. A, Candida albicans ATCC90029; C. M, Candida mycoderma; C. U, Candida utilis ATCC9950; A. F, Aspergillus flavus. A = Chloromycin; B = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSNorfloxacin; C = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole. |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3a | 256 | 512 | 256 | 256 | 256 | 128 | 512 | 512 | 256 | 256 | 256 | 128 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3b | 256 | 256 | 256 | 256 | 256 | 128 | 512 | 512 | 256 | 256 | 64 | 256 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3c | 128 | 128 | 64 | 128 | 128 | 64 | 256 | 512 | 128 | 256 | 128 | 128 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3d | 256 | 512 | 256 | 128 | 512 | 256 | 512 | 512 | 256 | 256 | 256 | 128 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound4a | 128 | 256 | 128 | 128 | 128 | 128 | 128 | 128 | 64 | 64 | 64 | 64 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound4b | 128 | 128 | 128 | 64 | 128 | 64 | 256 | 128 | 64 | 128 | 32 | 128 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound4c | 64 | 32 | 32 | 32 | 64 | 32 | 128 | 64 | 32 | 16 | 32 | 32 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound4d | 128 | 128 | 64 | 128 | 256 | 128 | 256 | 256 | 128 | 64 | 64 | 32 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5a | 256 | 256 | 128 | 256 | 256 | 128 | 256 | 256 | 128 | 128 | 64 | 256 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5b | 256 | 256 | 128 | 256 | 256 | 256 | 256 | 128 | 64 | 256 | 128 | 256 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5c | 128 | 128 | 256 | 256 | 512 | 128 | 128 | 512 | 128 | 128 | 128 | 128 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5d | 64 | 128 | 128 | 256 | 128 | 256 | 256 | 128 | 64 | 128 | 128 | 256 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5e | 128 | 256 | 256 | 256 | 256 | 64 | 128 | 256 | 128 | 512 | 256 | 256 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5f | 256 | 128 | 128 | 512 | 256 | 128 | 256 | 128 | 64 | 128 | 128 | 128 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5g | 128 | 128 | 256 | 256 | 128 | 256 | 128 | 128 | 128 | 64 | 64 | 128 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5h | 128 | 256 | 128 | 128 | 128 | 128 | 64 | 128 | 128 | 128 | 128 | 256 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5i | 128 | 128 | 512 | 512 | 512 | 256 | 256 | 256 | 64 | 64 | 512 | 128 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5j | 128 | 256 | 256 | 128 | 128 | 128 | 256 | 128 | 64 | 128 | 128 | 64 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5k | 256 | 256 | 128 | 128 | 256 | 256 | 128 | 256 | 128 | 128 | 256 | 128 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5l | 128 | 128 | 256 | 256 | 128 | 256 | 256 | 128 | 64 | 128 | 64 | 128 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6a | 32 | 64 | 32 | 64 | 32 | 16 | 32 | 64 | 16 | 8 | 8 | 16 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6b | 128 | 128 | 64 | 128 | 128 | 64 | 64 | 64 | 64 | 32 | 16 | 32 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6c | 64 | 64 | 32 | 64 | 64 | 64 | 64 | 128 | 32 | 32 | 16 | 64 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6d | 64 | 64 | 32 | 64 | 64 | 32 | 64 | 32 | 32 | 16 | 32 | 32 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6e | 64 | 32 | 64 | 64 | 64 | 64 | 32 | 64 | 64 | 16 | 16 | 32 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6f | 64 | 64 | 64 | 256 | 128 | 64 | 64 | 64 | 32 | 16 | 16 | 32 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 32 | 32 | 16 | 32 | 16 | 16 | 16 | 32 | 8 | 4 | 4 | 8 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6h | 64 | 128 | 32 | 64 | 32 | 32 | 64 | 64 | 16 | 32 | 16 | 32 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6i | 32 | 16 | 64 | 32 | 32 | 16 | 32 | 32 | 16 | 16 | 8 | 16 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6j | 64 | 64 | 64 | 64 | 64 | 64 | 32 | 64 | 32 | 32 | 16 | 32 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6k | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 64 | 32 | 16 | 16 | 32 |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6l | 32 | 64 | 32 | 32 | 32 | 32 | 32 | 32 | 16 | 8 | 8 | 16 |
| A | 8 | 16 | 32 | 8 | 32 | 16 | 16 | 32 | — | — | — | — |
| B | 4 | 1 | 2 | 1 | 4 | 4 | 1 | 2 | — | — | — | — |
| C | — | — | — | — | — | — | — | — | 1 | 4 | 8 | 256 |
The antifungal evaluation in vitro revealed that the activities of most of the compounds were relatively better in comparison to their antibacterial activities. The halobenzyl substituted target compounds 6a–l and alkyl substituted target compounds 4a–d exhibited moderate to excellent antifungal activities against the tested strains. Especially the target 2-chlorobenzyl compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g displayed comparable or even a stronger antifungal efficiency (MIC = 4–8 μg mL−1) against the tested fungi in comparison with the reference drug COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole (MIC = 1–256 mg mL−1). The length of the aliphatic chain also exhibited obvious effects on antifungal activity. The suitable length of the alkyl chain to exert the best antifungal efficacy was observed to be (CH2)9, and decyl tetrazole COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound4c gave generally better activity in comparison with other alkyl tetrazole compounds with shorter or longer chain length.
In addition, the combinations of tetrazole compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g with clinical antibacterials Chloromycin and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSNorfloxacin, or antifungal COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole respectively against the tested bacteria and fungi were investigated. To our excitement, the tested results showed excellent antimicrobial efficacies with less dosage and a broad antimicrobial spectrum as described in Tables 2 and 3. The FIC index was less than 0.8 which suggested that the combinations of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g with clinical drugs have good synergistic effects. As shown in Table 2, the combinations of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g with Chloromycin (4 μg mL−1) or COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSNorfloxacin (0.5 μg mL−1) were four- or two-fold more potent than themselves alone against MRSA. Interestingly, the combination of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole also displayed good activity against all the tested fungi with low MIC values of 2–4 μg mL−1 as described in Table 3. Especially, this combination showed excellent activity against COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole-insensitive Aspergillus flavus in comparison with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole alone (MIC = 256 μg mL−1). These results manifested that the combinations of tetrazoles with clinical drugs could efficiently enhance antimicrobial activity, overcome drug resistance and broaden the antimicrobial spectrum. The advantages of combinations might be attributed to the different binding sites of these compounds towards the tested microorganism.
| Bacteria | Compds | MIC | FIC index | Effect | Compds | MIC | FIC index | Effect |
Minimum inhibitory concentrations were determined by micro broth dilution method for microdilution plates. MRSA, Methicillinresistant Staphylococcus aureus N315; S. A, Staphylococcus aureus ATCC25923; B. S, Bacillus subtilis ATCC6633; M. L, Micrococcus luteus ATCC 4698; E. C, Escherichia coli DH52; S. D, Shigella dysenteriae ATCC51252; P. A, Pseudomonas aeruginosa ATCC27853; B. P, Bacillus proteus ATCC13315; C. A, Candida albicans ATCC90029; C. M, Candida mycoderma; C. U, Candida utilis ATCC9950; A. F, Aspergillus flavus. A = Chloromycin; B = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSNorfloxacin; C = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole. |
| S. A | A | 4 | 0.750 | Synergistic | B | 2 | 0.500 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 16 | COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 16 |
| MRSA | A | 4 | 0.500 | Synergistic | B | 0.5 | 0.750 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 8 | COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 8 |
| B. S | A | 4 | 0.375 | Synergistic | B | 1 | 0.625 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 16 | COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 8 |
| M. L | A | 2 | 0.500 | Synergistic | B | 0.5 | 0.750 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 8 | COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 8 |
| B. P | A | 8 | 0.750 | Synergistic | B | 1 | 0.500 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 32 | COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 16 |
| E. C | A | 8 | 0.750 | Synergistic | B | 0.5 | 0.625 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 16 | COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 8 |
| P. A | A | 4 | 0.500 | Synergistic | B | 0.5 | 0.750 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 16 | COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 16 |
| S. D | A | 8 | 0.500 | Synergistic | B | 0.5 | 0.625 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 8 | COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 4 |
| Fungi | Compds | MIC (μg mL−1) | FIC index | Effect |
Minimum inhibitory concentrations were determined by micro broth dilution method for microdilution plates. MRSA, Methicillinresistant Staphylococcus aureus N315; S. A, Staphylococcus aureus ATCC25923; B. S, Bacillus subtilis ATCC6633; M. L, Micrococcus luteus ATCC 4698; E. C, Escherichia coli DH52; S. D, Shigella dysenteriae ATCC51252; P. A, Pseudomonas aeruginosa ATCC27853; B. P, Bacillus proteus ATCC13315; C. A, Candida albicans ATCC90029; C. M, Candida mycoderma; C. U, Candida utilis ATCC9950; A. F, Aspergillus flavus. A = Chloromycin; B = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSNorfloxacin; C = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole. |
| C. A | C | 0.5 | 0.750 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 2 |
| C. M | C | 1 | 0.500 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 2 |
| C. U | C | 2 | 0.500 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 4 |
| A. F | C | 64 | 0.500 | Synergistic |
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g | 4 |
3.2 Values of ionization constant (pKa) and lg P
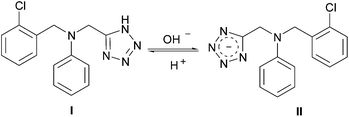
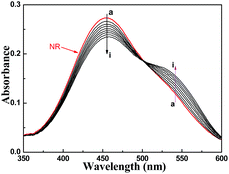
The ionization constant (pKa) of a molecule is an important physicochemical parameter which has significant influence on its lipophilicity, solubility, protein binding as well as permeability. Therefore the pKa values are usually used to evaluate the extent of ionization of bioactive molecules at different pH values, which is of fundamental importance in the consideration of their pharmacokinetic behavior such as absorption, distribution, metabolism, and excretion in organisms.37,38 A compound with a pKa < 4 or > 10 will be charged at physiological pH and display slower diffusion rate across biological membranes such as the blood–brain barrier. UV spectroscopy is an efficient method for the determination of ionization constants which employs readily available equipment with a fast determination. Structural transposition of title tetrazole compounds would be generated in buffer solutions at different pH values, thus resulting in the changes in UV absorbance. The changes observed in the UV spectrum of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g in different buffer solutions are clearly displayed in Fig. 1. With the pH values ranging from 2.0 to 8.5, the structural formula of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g was changed from COMPOUND LINKS
Read more about this on ChemSpider
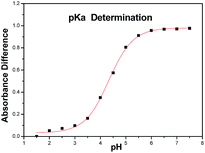
Download mol file of compoundI to II (Scheme 2). As shown in Fig. 2, the spectral differences between 283 and 309 nm were chosen to analyze the pKa of target tetrazole compounds. Data analysis included normalization of the raw scans (Abs330 nm = 0), and then the spectral difference between the acid spectra and the spectra obtained at every other pH was calculated. The wavelengths of maximum positive and negative deviations were determined graphically, and the absolute values of the absorbance difference at the chosen wavelengths were summed. The total absorbance difference was then plotted versus pH, and the data were fit to eqn (1)| |  | (1) |
where εHA and εA− are the extinction coefficients of the acid and base forms of the compound, respectively, and [St] is the total compound concentration. When using absorbance differences, εHA and εA− are simply the minima and maxima of the curve.39 As shown in Table 4, the pKa values of compounds 4a–d and 6a–l displayed an appropriate range 4.13–4.83, which suggested their good pharmacokinetic behavior as bioactive agents.
Table 4 pKa, lg P and c lg P of target compounds 4a–d and 6a–la
Lipophilicity/hydrophilicity plays a significant role in determining where drugs are distributed and how rapidly they are metabolized and excreted in the body, for example, hydrophobic drugs are preferentially distributed to hydrophobic compartments such as lipid bilayers of cells while hydrophilic drugs are preferentially found in hydrophilic compartments like blood serum.40 Therefore, the lipophilicity/hydrophilicity expressed as lg P was calculated theoretically using ChemDraw Ultra 10.0 software and experimentally by a traditional saturation shake flask approach combined with a UV-vis spectrophotometric method. The obtained results are given in Table 4, the c lg P of compounds 4a–d increased with the increase in the length of the alkyl chain, and an enhancement of the antimicrobial activities was observed in compounds 4a–c, but decreased in that of compounds 4c–d, these might be explained by the possibility that higher lipophilic compounds were unfavourable for being delivered to the binding sites. This phenomenon also indicted that suitable lipophilicity is necessary for good activities in drug design (Fig. 3).
3.3 Interaction with calf thymus DNA
3.3.1 UV-vis absorption spectral study. The application of UV-vis absorption measurement is one of the most important techniques in DNA-binding studies. Generally, hypochromism and hyperchromism are important spectral features to distinguish the change of DNA double-helical structure. The hyperchromism originates from the breakage of the DNA duplex secondary structure; while the hypochromism generates from the stabilization of the DNA duplex by either the intercalation binding mode or the electrostatic effect of small molecules.41 The hypochromism demonstrated a close proximity of the aromatic chromophore to the DNA bases, which might be attributed to the strong interaction between the electronic states of the intercalating chromophore and that of the DNA base. With a fixed concentration of DNA, UV-vis absorption spectra were recorded with the increasing amount of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g. As shown in Fig. 4, UV-vis spectra showed that the maximum absorption of DNA at 260 nm displayed a proportional increase with the increasing concentration of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g. Simultaneously, the absorption value of the sum of free DNA and free compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g was a little greater than the measured value of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g–DNA complex, which was observed in the inset of Fig. 4. These indicated that a weak hypochromic effect existed between DNA and compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g. Moreover, the intercalation of the aromatic chromophore of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g into the helix and the strong overlap of π–π* states in the large π-conjugated system with the electronic states of DNA bases were consistent with the observed spectral changes.42,43 On the basis of the variations in the absorption spectra of DNA upon binding to tetrazole derivative COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g, eqn (2) can be utilized to calculate the binding constant (K).
| |  | (2) |
A 0 and A represent the absorbance of DNA in the absence and presence of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g at 260 nm, ξC and ξD−C are the absorption coefficients of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g–DNA complex respectively. The plot of A0 (A−1−A0) versus 1/[compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g] is constructed by using the absorption titration data and linear fitting (Fig. 5), yielding the binding constant, K = 4.16 × 104 L mol−1, R = 0.999, and SD = 0.17 (R is the correlation coefficient. SD is standard deviation).
4. Conclusions
In conclusion, a novel type of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTStetrazole was successfully synthesized for the first time via an easy, convenient and efficient synthetic procedure starting from commercially available substituted anilines. The antimicrobial tests demonstrated that some prepared compounds exhibited comparable or even superior antifungal activity against the tested strains relative to the reference drug COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole. Especially compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g was found to be four-fold more potent than COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole against Aspergillus flavus (MIC = 256 μg mL−1). Meanwhile, the combinations of tetrazoles with antibacterials Chloromycin and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSNorfloxacin or antifungal COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole gave striking enhanced antimicrobial efficiency with less dosage as well as a broader antimicrobial spectrum than their separate use. Notably, the combined system was more sensitive to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSFluconazole-insensitive Aspergillus flavus and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTSmethicillin-resistant MRSA. The specific interaction of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g with calf thymus DNA, which was studied by UV-vis absorption spectroscopy, manifested that compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g could intercalate into DNA to form the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound6g–DNA complex which might further block DNA replication to exert its good antibacterial and antifungal activities. All these results should be a promising starting point to optimize the structures of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound
Explore further on Open PHACTStetrazole-derived tertiary amines to access potent antimicrobial agents. Acknowledgements
This work was partially supported by the National Natural Science Foundation of China [no. 21172181, 21372186, 81350110523, 81450110095 (The Research Fellowship for International Young Scientists from International (Regional) Cooperation and Exchange Program)], the key program from the Natural Science Foundation of Chongqing (CSTC2012jjB10026), the Specialized Research Fund for the Doctoral Program of Higher Education of China (SRFDP 20110182110007) and Chongqing Special Fund for Postdoctoral Research Proposal (Xm2014127). References
- R. J. Scheffler, S. Colmer, H. Tynan, A. L. Demain and V. P. Gullo, Appl. Microbiol. Biotechnol., 2013, 97, 969–976 CrossRef CAS PubMed.
- L. Zhang, X. M. Peng, G. L. V. Damu, R. X. Geng and C. H. Zhou, Med. Res. Rev., 2014, 34, 340–437 CrossRef CAS PubMed.
- X. M. Peng, G. X. Cai and C. H. Zhou, Curr. Top. Med. Chem., 2013, 13, 1963–2010 CrossRef CAS.
- X. M. Peng, G. L. V. Damu and C. H. Zhou, Curr. Pharm. Des., 2013, 19, 3884–3930 CrossRef CAS.
- Y. Hokai, B. Jurkowicz, J. Fernández-Gallardo, N. Zakirkhodjaev, M. Sanaú, T. R. Muth and M. Contel, J. Inorg. Biochem., 2014, 138, 81–88 CrossRef CAS PubMed.
- M. I. Cassetta, T. Marzo, S. Fallani, A. Novelli and L. Messori, Biometals, 2014, 27, 787–791 CrossRef CAS PubMed.
- N. Petrosillo, M. Giannella, R. Lewis and P. Viale, Expert Rev. Anti-Infect. Ther., 2013, 11, 159–177 CrossRef CAS PubMed.
- J. J. Vehreschild, A. Birtel, M. J. G. T. Vehreschild, B. Liss, F. Farowski, M. Kochanek, M. Sieniawski, A. Steinbach, K. Wahlers and G. Faetkenheuer, Crit. Rev. Microbiol., 2013, 39, 310–324 CrossRef CAS PubMed.
- S. F. Cui, Y. Wang, C. H. Zhou and R. X. Geng, Sci. Sin.: Chim., 2012, 42, 1105–1131 CrossRef CAS.
- S. F. Cui, Y. Ren, S. L. Zhang, X. M. Peng, G. L. V. Damu, R. X. Geng and C. H. Zhou, Bioorg. Med. Chem. Lett., 2013, 23, 3267–3272 CrossRef CAS PubMed.
- B. T. Yin, C. Y. Yan, X. M. Peng, S. L. Zhang, S. Rasheed, R. X. Geng and C. H. Zhou, Eur. J. Med. Chem., 2014, 71, 148–159 CrossRef CAS PubMed.
- L. L. Dai, S. F. Cui, G. L. V. Damu and C. H. Zhou, Chin. J. Org. Chem., 2013, 33, 224–244 CrossRef CAS.
- J. Roh, K. Vavrova and A. Hrabalek, Eur. J. Org. Chem., 2012, 2012, 6101–6118 CrossRef CAS.
- O. Mesenzani, A. Massarotti, M. Giustiniano, T. Pirali, V. Bevilacqua, A. Caldarelli, P. Canonico, G. Sorba, E. Novellino, A. A. Genazzani and G. C. Tron, Bioorg. Med. Chem. Lett., 2011, 21, 764–768 CrossRef CAS PubMed.
- R. Romagnoli, P. G. Baraldi, M. K. Salvador, D. Preti, M. A. Tabrizi, A. Brancale, X. H. Fu, J. Li, S. Z. Zhang, E. Hamel, R. Bortolozzi, G. Basso and G. Viola, J. Med. Chem., 2012, 55, 475–488 CrossRef CAS PubMed.
- K. Pegklidou, C. Koukoulitsa, I. Nicolaou and V. J. Demopoulos, Bioorg. Med. Chem., 2010, 18, 2107–2114 CrossRef CAS PubMed.
- C. F. Matta, A. A. Arabi and D. F. Weaver, Eur. J. Med. Chem., 2010, 45, 1868–1872 CrossRef CAS PubMed.
- B. J. Al-Hourani, S. K. Sharma, J. Y. Mane, J. Tuszynski, V. Baracos, T. Kniess, M. Suresh, J. Pietzsch and F. Wuest, Bioorg. Med. Chem. Lett., 2011, 21, 1823–1826 CrossRef CAS PubMed.
- W. A. Carroll, D. M. Kalvin, A. P. Medrano, A. S. Florjancic, Y. Wang, D. L. Donnelly-Roberts, M. T. Namovic, G. Grayson, P. Honoréa and M. F. Jarvis, Bioorg. Med. Chem. Lett., 2007, 17, 4044–4048 CrossRef CAS PubMed.
- Y. Shi, C. H. Zhou, X. D. Zhou, R. X. Geng and Q. G. Ji, Acta Pharmacol. Sin., 2011, 46, 798–810 CAS.
- Y. Y. Zhang and C. H. Zhou, Bioorg. Med. Chem. Lett., 2011, 21, 4349–4352 CrossRef CAS PubMed.
- S. L. Zhang, G. L. V. Damu, L. Zhang, R. X. Geng and C. H. Zhou, Eur. J. Med. Chem., 2012, 55, 164–175 CrossRef CAS PubMed.
- F. F. Zhang, L. L. Gan and C. H. Zhou, Bioorg. Med. Chem. Lett., 2010, 20, 1881–1184 CrossRef CAS PubMed.
- R. J. MacFadyen, M. Tree, A. F. Lever and J. L. Reid, Clin. Sci., 1992, 83, 549–556 CAS.
- M. Karaman, S. Balta, S. A. Ay, M. Cakar, I. Naharci, S. Demirkol, T. Celik, Z. Arslan, O. Kurt, N. Kocak, H. Sarlak, S. Demirbas, F. Bulucu and E. Bozoglu, Clin. Exp. Hypertens., 2013, 35, 516–522 CrossRef CAS PubMed.
- C. H. Lee, J. W. Liu, C. C. Li, C. C. Chien, Y. F. Tang and L. H. Su, Antimicrob. Agents Chemother., 2011, 55, 4058–4063 CrossRef CAS PubMed.
- L. Pochini, M. Galluccio, D. Scumaci, N. Giangregorio, A. Tonazzi, F. Palmieri and C. Indiveri, Chem.-Biol. Interact., 2008, 173, 187–194 CrossRef CAS PubMed.
- N. Sigmond, M. Baechtold, P. M. Schumacher, V. Hartwich, T. W. Schnider and M. Luginbuehl, Br. J. Anaesth., 2013, 111, 197–208 CrossRef CAS PubMed.
- A. T. Nguyen-Trung, D. Tritsch, C. Grosdemange-Billiard and M. Rohmer, Bioorg. Med. Chem. Lett., 2013, 23, 1643–1647 CrossRef CAS PubMed.
- G. H. Vondenhoff, B. Gadakh, K. Severinov and A. Van Aerschot, ChemBioChem, 2012, 13, 1959–1969 CrossRef CAS PubMed.
- C. H. Zhou and Y. Wang, Curr. Med. Chem., 2012, 19, 239–280 CrossRef CAS.
- Y. Wang, G. L. V. Damu, J. S. Lv, R. X. Geng, D. C. Yang and C. H. Zhou, Bioorg. Med. Chem. Lett., 2012, 22, 5363–5366 CrossRef CAS PubMed.
- V. S. Pore, M. A. Jagtap, S. G. Agalave, A. K. Pandey, M. S. Siddiqi, V. Kumar and P. K. Shukla, MedChemComm, 2012, 3, 484–488 RSC.
- B. Fang, C. H. Zhou and X. C. Rao, Eur. J. Med. Chem., 2010, 45, 4388–4398 CrossRef CAS PubMed.
- S. L. Zhang, J. J. Chang, G. L. V. Damu, R. X. Geng and C. H. Zhou, MedChemComm, 2013, 4, 839–846 RSC.
- H. Z. Zhang, G. L. V. Damu, G. X. Cai and C. H. Zhou, Eur. J. Med. Chem., 2013, 64, 329–344 CrossRef CAS PubMed.
- D. T. Manallack, Perspect. Med. Chem., 2007, 1, 25–38 Search PubMed.
- R. I. Allen, K. J. Box, J. E. A. Comer, C. Peake and K. Y. Tam, J. Pharm. Biomed. Anal., 1998, 17, 699–712 CrossRef CAS.
- S. F. Cui, L. P. Peng, H. Z. Zhang, S. Rasheed, K. Vijaya Kumar and C. H Zhou, Eur. J. Med. Chem., 2014, 86, 318–334 CrossRef CAS PubMed.
- L. Zhang, J. J. Chang, S. L. Zhang, G. L. V. Damu, R. X. Geng and C. H. Zhou, Bioorg. Med. Chem., 2013, 21, 4158–4169 CrossRef CAS PubMed.
- M. Rahban, A. Divsalar, A. A. Saboury and A. Golestani, J. Phys. Chem. C, 2010, 114, 5798–5803 CAS.
- G. W. Zhang, P. Fu, L. Wang and M. M. Hu, J. Agric. Food Chem., 2011, 59, 8944–8952 CrossRef CAS PubMed.
- X. L. Li, Y. J. Hu, H. Wang, B. Q. Yu and H. L. Yue, Biomacromolecules, 2012, 13, 873–880 CrossRef CAS PubMed.
- Y. Ni, S. Du and S. Kokot, Anal. Chim. Acta, 2007, 584, 19–27 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4md00266k |
| ‡ Postdoctoral fellow from School of Chemistry, Madurai Kamaraj University, India. |
| § Postdoctoral fellow from Department of Chemistry, University of Hyderabad, India. |
|
| This journal is © The Royal Society of Chemistry 2015 |
Click here to see how this site uses Cookies. View our privacy policy here.