Open Access Article
Open Access ArticleNMR-based investigation of the Drosophila melanogaster metabolome under the influence of daily cycles of light and temperature†
Navdeep
Gogna‡
a,
Viveka Jagdish
Singh‡
b,
Vasu
Sheeba
 b and
Kavita
Dorai
*a
b and
Kavita
Dorai
*a
aIndian Institute of Science Education & Research (IISER) Mohali, Knowledge City Sector 81, Mohali PO, Manauli 140306, Punjab, India. E-mail: kavita@iisermohali.ac.in; Tel: +91-0172-2240266
bJawaharlal Nehru Centre for Advanced Scientific Research, Jakkur P.O., Bengaluru, 560064 Karnataka, India. Tel: +91-080-2208-2580
First published on 21st September 2015
Abstract
We utilized an NMR-based metabolomic approach to profile the metabolites in Drosophila melanogaster that cycle with a daily rhythm. 1H 1D and 2D NMR experiments were performed on whole-body extracts sampled from flies that experienced strong time cues in the form of both light and temperature cycles. Multivariate and univariate statistical analysis was used to identify those metabolites whose concentrations oscillate diurnally. We compared metabolite levels at two time points twelve hours apart, one close to the end of the day and the other close to the end of the night, and identified metabolites that differed significantly in their relative concentrations. We were able to identify 14 such metabolites whose concentrations differed significantly between the two time points. The concentrations of metabolites such as sterols, fatty acids, amino acids such as leucine, valine, isoleucine, alanine and lysine as well as other metabolites such as creatine, glucose, AMP and NAD were higher close to the end of the night, whereas the levels of lactic acid, and a few amino acids such as histidine and tryptophan were higher close to the end of the day. We compared signal intensities across 12 equally spaced time points for these 14 metabolites, in order to profile the changes in their levels across the day, since the NMR metabolite peak intensity is directly proportional to its molar concentration. Through this report we establish NMR-based metabolomics combined with multivariate statistical analysis as a useful method for future studies on the interactions between circadian clocks and metabolic processes.
Introduction
It has become increasingly critical to reconcile with the fact that metabolic processes are subject to cyclic influences not only from the external environment but also from the internal circadian clock. It can be argued that homeostatic processes that involve metabolic pathways also feedback to the circadian clock. Today's lifestyle often places harsh demands on human physiology that is not always in-sync with the evolutionary forces that may have shaped circadian clocks and our metabolic processes. This may in part, have a role to play in the rapid rise in metabolic disorders in modern human populations across the world.Recently several studies point to the intricate connections between circadian clocks and metabolic pathways involved in fatty acid or glucose metabolism in the liver.1 It is likely that intrinsic adaptive advantages are offered by circadian clock mediation of metabolic processes, in being able to provide organisms with an efficient way to deal with toxic metabolic wastes and, most effectively utilise any unstable intermediate in these pathways due to the anticipatory ability of time-keeping machinery. Circadian clocks regulate the behavioural process of feeding, which may in turn affect metabolic processes.2,3 Since feeding itself is rhythmic in many organisms, it also directly causes oscillations in metabolic processes. The molecules NAD+, SIRT1 and AMP Kinase (AMPK) are known to be sensors of the nutritional or energy status of cells.4–6 SIRT1 deacetylates several proteins involved in varied processes such as gluconeogenesis and cholesterol metabolism,4 thus converting the information on cyclic nutritional status to activate appropriate metabolic processes. AMPK also interacts with the circadian clock by acting on NAD+ levels.6 Thus, a complex intertwining of processes in these apparently distinct entities namely circadian clocks, feeding-related behaviours and the metabolic machinery of organisms remains to be unraveled.
Studies on mammals have shown that animals with mutations in circadian clock genes often develop obesity, metabolic syndrome, hypoinsulinaemia and diabetes,7,8 all of which are indicative of major disruptions in multiple metabolic pathways. Furthermore, several studies of rhythmically expressed transcripts in the whole genome have suggested that genes involved in lipid, glucose and fatty acid metabolism are regulated in a rhythmic manner by a few circadian clock genes.9–11 Such relationships have also been shown to exist in fruit flies Drosophila melanogaster,9,12 where the circadian clocks in the fat body (tissue equivalent to the liver) control cyclic expression of several genes. While genetic amenability of fruit flies offers the possibility of highly targeted tissue specific manipulation,13 yet, there have been relatively fewer studies that address the nature of the relationship between circadian clocks, metabolic processes and feeding behaviours in flies. This may be attributed to the fact that feeding cannot be as easily measured or manipulated in flies. However, studies by Amita Sehgal and colleagues have shown that ClockJrk mutation which disrupts activity/rest rhythm also changes the peak of feeding.14 This study revealed that tissue specific disruption of the clock was limited to the fat body also causing several behavioural and physiological defects. Such flies are unable to exhibit cyclic feeding, and show overall higher feeding, especially at night, yet such flies are more prone to starvation and have reduced glycogen stores. On the other hand, when disruption of circadian clocks is restricted to the central clock neurons, then flies show increased starvation resistance and glycogen storage. The same group also went on to show that similar to mammals, circadian clocks in the fat body control cyclic expression of several genes suggesting that metabolism is likely to be regulated via transcriptional control by a few circadian genes.12 Thus the study provides evidence for links between organs known to be important for metabolic processes and circadian clocks in flies. While the studies discussed above used cycling transcripts as readouts for rhythmic metabolic processes, an alternate and more recent approach has been used to estimate levels of metabolites themselves. This is because even in the absence of cycling transcripts it is possible to envisage oscillations in metabolic processes based on post transcriptional/translational modifications leading to rhythmicity in the activation state of enzymes and their substrates.15–19 Such enzymes, when involved in the production, activation, deactivation or quenching of metabolites can also generate rhythmicity in metabolite levels. An example of a study that examined metabolites includes one in which excessive weight gain was prevented in mice under a regime of night-restricted high-fat diet (which under ad libitum conditions induces obesity). Metabolite analysis showed that both fatty acid and glucose metabolism that were altered by ad libitum high fat food could be restored to normal by appropriate timing of feeding (mice being nocturnal feeders). Interestingly, small molecules such as NAD+, FAD, cADPR are known to oscillate in a circadian manner.1,18,20,21 Studies in mice have shown that NAD+ feeds back to the circadian clock through the activation of a histone deacetylase enzyme Sirtuin1 (SIRT1).16,21–23 These studies suggest that metabolites as readouts can be a more direct approach to study the link between circadian clocks and metabolism.
Metabolomics seeks to obtain the complete metabolic profile of an organism. NMR spectroscopy and mass spectrometry are two analytical techniques widely used for metabolomic studies.24,25 Recently, in an attempt to develop a ‘metabolite timetable’ from blood plasma which could potentially act as a diagnostic tool, Minami and colleagues analysed the blood metabolome of mice by LC/MS to differentiate between a large number of metabolites.26 One study which assayed a large number of liver metabolites of mice using GC/MS or LC/MS clearly demonstrates the critical role of timing of feeding and the underlying metabolic processes.27 A separate study by Eckel-Mahan and colleagues analysed the liver metabolome of mice under LD cycles by LC/MS and unravelled a large number of cycling metabolites of which, those involved in lipid, carbohydrate and nucleotide metabolism peaked during the day which is the time when mice are resting, while those involved in amino acid metabolism and processing of xenobiotics peaked during night, which is their active phase.28 In addition to these, three separate groups reported results from analysis of the metabolome of humans from saliva and/or blood plasma by LC/GC MS.29–31 However, due to limited molecular weight distribution of the metabolites, mass spectrometry usually requires chromatographic separation, which inevitably disturbs the relative concentration of the metabolites and hence the metabolome being characterized. Mass spectrometry is also restricted to those metabolites that ionize readily; molecules such as carbohydrates do not ionize well. In the wake of such limitations, NMR is preferred over mass spectrometry for such metabolites.
High-resolution NMR spectroscopy is a non-destructive, quantitative technique to obtain metabolite concentrations with the advantages of high-reproducibility, rapid data analysis minimal sample preparation. Metabolites are identified by their characteristic resonance peak positions and spectral patterns and are usually validated by comparison with NMR spectra of pure compounds deposited in standard databases. Previously, low sensitivity and peak overlap problems of NMR limited its use in metabolomics, however, recent advantages in NMR instrumentation such as higher field magnets, isotope labeling and cryoprobes have led to its widespread use as a metabolomics tool. NMR metabolomics is able to simultaneously measure the variation in relative concentrations of a large number of metabolites and the peak intensity in an NMR spectrum is directly proportional to the concentration of the metabolite.
Most NMR-based metabolomic investigations of Drosophila melanogaster have focused on organismal response to different environmental stresses.32–40 However, only a handful of studies have tried to directly estimate the daily profile of metabolites within tissues while most others have relied on the oscillation in gene transcripts as indicators of rhythmicity in metabolic function. While the fly has been a workhorse for circadian rhythm research for several decades, only recently has there been a move towards using the fly to study links between clocks and metabolism. Overall, these studies suggest interlinking of circadian clocks and energy homeostatic processes although the nature of such interactions is far from clear. In flies it has been demonstrated that circadian clock in peripheral tissues are sensitive to daily cycles of food availability12 and that starvation can disrupt the circadian clock-modulated sleep/wake behaviour.41 To our knowledge, all previous studies on flies have relied on either gene transcripts or the analysis of a few well studied metabolites to estimate metabolic function.12,14,42,43 The work of the Panda, Ueda and Sassone Corsi labs on rodents have shown that large scale analysis of metabolites can provide much greater information regarding the interactions between metabolic pathways and the clock.15,26–28 Hence our study is aimed to perform the first step of rigorously cataloguing the profile of rhythmic metabolites in the fly when subjected to a 12:12 h day:night cycle with light and warm temperature during the day and darkness and cool temperature at night. Since this study was intended to establish that this method would indeed recognize metabolite cycling, we used a regime that would ensure robust cycling in physiological processes. Since both light and temperature are known to be strong time cues and act synergistically on the circadian clock we adopted the above protocol.44 This would allow us to identify those metabolites that cycle with a daily rhythm, although it would also include those that are rhythmic merely as a response to cyclic light and/or temperature levels. We performed untargeted metabolite profiling of D. melanogaster by 1D 1H and 2D NMR spectroscopy in conjunction with OPLS-DA pattern recognition analysis. We were able to identify several metabolites whose concentrations undergo diurnal changes in the presence of cyclic time cues in the form of light and temperature. Thus, we establish the validity of this approach in performing quantitative measurements of cycling metabolites in the fly, which is rapidly becoming a very useful model system in the area of metabolomics.36,38
Results and discussion
Metabolite identification by 1D and 2D NMR
A detailed analysis of the metabolite profile of D. melanogaster was performed using 1D 1H and 2D NMR experiments. The resonances were assigned to specific metabolites based on their chemical shift values matching with the values of the pure standard compounds extracted from spectral databases such as MMCD and BMRB. A variety of metabolites were identified on the basis of their chemical shift values and scalar coupling patterns (ESI†). We also confirmed the metabolite identification using unambiguous signal assignments from two-dimensional NMR spectra, including homonuclear experiments such as COSY and TOCSY, and heteronuclear experiments such as HSQC and HMQC.Multivariate statistical analysis
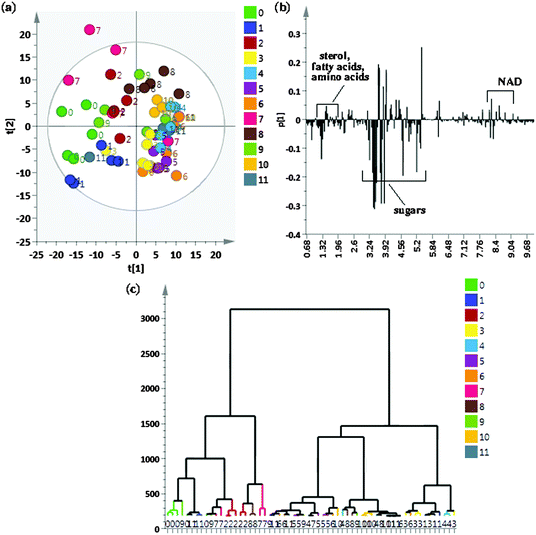
To identify metabolites that show cycling in concentrations after exposure to a 12:12 hour, light:dark (LD) + warm (28 °C):cold (18 °C) cycles, flies were sampled at 12 time points across a 24 h period. We began by an initial comparison of samples from all 12 time points using the unsupervised method of PCA to look for possible outliers, groups, similarities and other patterns in the data. Fig. 1(a) shows the PCA score plots for all time points taken together with component 1 explaining 30.5% of the variation and component 2 explaining 24.6% of the variation. As can be seen, replicates of individual time points cluster well however there are overlaps between different time points resulting in no clear separation when all the time points were taken together. As can be seen from the PCA score plot, component 1 is able to separate ZT22, ZT20 and ZT18 (0, 1 and 2 respectively) time points from the rest of the time points. ZT8 (7) replicates did not show proper clustering and had outliers and ZT6 (8) samples could also be separately seen from rest of the time point samples. Fig. 1(b) shows the loading plot corresponding to the scores plot, showing the variables responsible for the cluster separation. We used hierarchical cluster analysis (HCA) to identify natural groupings in the data for all twelve time points. Fig. 1(c) shows the tree dendrogram prepared using HCA, which summarizes all the variation present in the dataset. No clear separation could be seen, although replicates within individual time points cluster fairly well both in the PCA score plot and in the twelve time point dendrogram. With R2 = 0.883 and Q2 = 0.767, the PCA model shows good predictability. Variables identified using PCA were subjected to ANOVA and p < 0.01 was considered to indicate a statistically significant difference. Table S2 (ESI†) shows metabolites identified using ANOVA that contribute to the separation and post hoc analysis to show which time points are significantly different.For better analysis and to obtain a clear picture for metabolite cycling, instead of taking individual replicates per time point, we considered the mean of all replicates per time point (which left us with 12 data points instead of 60 (12 × 5 = 60) corresponding to each time point sampled). In order to better visualize the results of the cluster analysis, we used a polar dendrogram, which maps the leaf nodes in a radial fashion around the circumference of a circle. Fig. 2 shows the polar dendrogram plotted for all twelve time points. As can be seen, three main nodes resulting in three groups can be identified in the dendrogram. Two groups have five time points each-one group has time points corresponding to the light phase (ZT4 to ZT12) whereas the other group has time points corresponding to the dark phase (ZT14 to ZT22). This shows a clear separation between metabolism during light and dark phases. A third separation shows two time points ZT0 and ZT2 as being different from both the other groups and falling in-between both the other groups, indicating that both these time points are different yet intermediate to both the groups. This might be the region of change in metabolism from day to night phases. Since the two groups separated have time points mostly 10–12 hours apart hinting at a possible 10–12 hour cycling in metabolism, we selected two time points ZT10 and ZT22, which were twelve hours apart, to proceed further with the analysis.
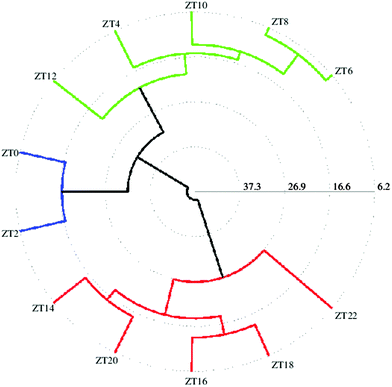 | ||
| Fig. 2 Similarities between different time points (from ZT0 to ZT22) represented as a polar dendrogram. All time points have five replicates each. | ||
OPLS-DA analysis
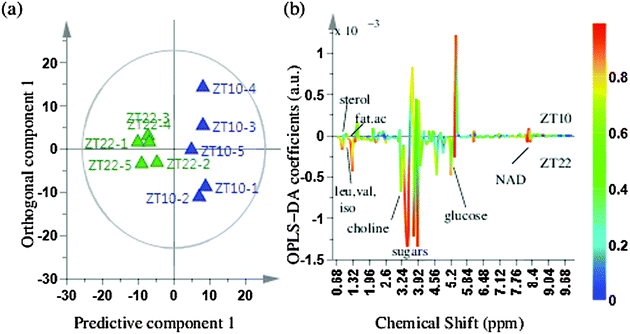
The PCA analysis showed some class differentiation, but there were noticeable overlaps. To address this problem, we used an OPLS-DA statistical approach. OPLS-DA can be used to maximize covariance between the measured data (NMR peak intensities) and the response variable (predictive classifications) and the OPLS-DA color-coded coefficient loading plot can be used to identify significant metabolites contributing to the separation between the two time points. A quantitative comparative analysis of NMR metabolomic profiles has to account for peak shifts across different spectra that could be caused by pH or temperature fluctuations or magnetic field inhomogeneity.45 Prior to the OPLS-DA analysis, we used the icoshift algorithm for alignment of all 1D NMR spectra, which is based on correlation shifting of spectral intervals, uses the FFT correlation for simultaneous spectral alignment, and avoids down-sampling steps such as binning.46Fig. 3(a) depicts the OPLS-DA score plot for the comparison between time points ZT10 and ZT22, obtained with one predictive component and one orthogonal component, showing a clear separation between the two time points. The loading plot shows the metabolites responsible for separation between the two time points (Fig. 3(b)). The VIP score parameter was used to confirm the variables identified to be responsible for time point separations. The variance explained by the model R2X was 0.957 and the variance predicted Q2 was 0.849. The model therefore was an effective model, with a good predictive accuracy. The model was also statistically significant after testing with CV-ANOVA with a p-value < 0.01 and was validated by a permutation test (p-value < 0.05) which proved the credibility and robust nature of this model. In addition to multivariate analysis, univariate analysis was also performed. The variables were tested for statistical significance using a t-test. The metabolite peaks for sterols including cortisol, fatty acid terminal methyl and fatty acid (CH2)n, amino acids like leucine, isoleucine, valine, alanine, lysine, other metabolites like AMP, NAD, creatine and cortisol were present in higher concentrations at ZT22 whereas metabolite peaks for histidine, tryptophan and lactic acid were present in lower concentrations at ZT22. A multiple hypothesis test correction method of Benjamini–Hochberg was also applied to confirm the statistical significance of metabolites identified, which further proved the statistical significance of the metabolites identified.As suggested by the polar dendrogram of twelve time points (Fig. 2), the separation occurs mainly in day and night phases which are 10–12 hours apart. We hence performed a further comparative analysis of time points which were 10–12 hours apart, in order to identify other metabolites with significant concentration changes that occur at other time points relative to ZT10 or ZT22. In addition to comparing the ZT10 and ZT22 time points (Fig. 3), we also compared ZT8 with ZT22, ZT6 with ZT18 and ZT4 with ZT16 (ESI†). Since ZT0 and ZT2 occur at the beginning of the day and ZT22 occurs towards the end of the night, we also compared metabolites between ZT0, ZT2 with ZT22 even though they are separated by only 5 hours, to detect metabolites that actively undergo changes during the transition from day to night. The comparison between ZT8 and ZT20 did not lead to the emergence of hitherto undiscovered cycling metabolites and the results matched with the results obtained for the comparison between ZT10 and ZT22. The comparison between ZT4 and ZT16 showed a lipid peak as identified by the terminal methyl peak, sugars like sucrose and erythrose and amino acids such as valine and leucine to be responsible for separation. The comparison between ZT6 and ZT18 showed peaks from amino acids valine, leucine and isoleucine and NAD to be responsible for separation. Comparison between ZT0 and ZT22 showed the same aliphatic amino acids and NAD to be involved in group separation whereas comparison between ZT2 and ZT22 showed additional metabolites of lipids and lysine to be contributing towards separation. Score plots for the above five time point comparisons, along with metabolites identified using the t-test (p < 0.01) to be responsible for group separations, are shown in the ESI.†
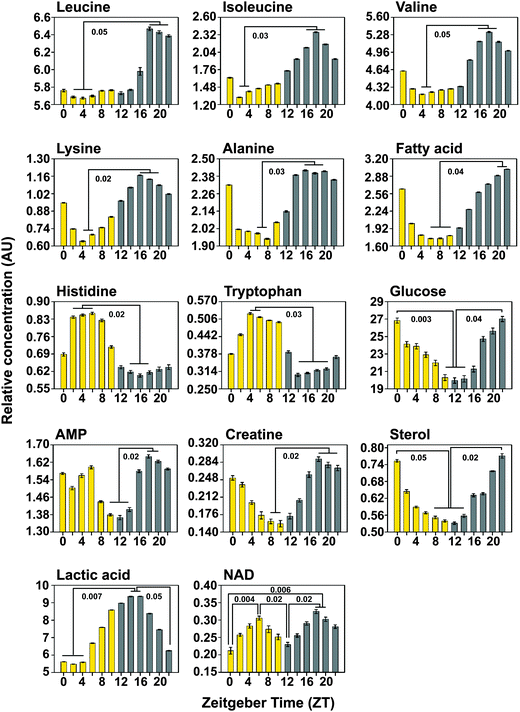
Variation in significant metabolites
Changes in metabolite concentrations over the 24 h LD warm:cold cycles were analyzed by: first, finding the spectral regions that changed significantly (p < 0.01) at two time points compared; second, the intensities of individual metabolites were assessed as integrals of the individual spectral peaks in those bins. Table 1 shows relative integrals of metabolites (at time points ZT10 and ZT22) that were found to be statistically different in concentrations at both these time points. To further confirm that these metabolites indeed show cycling with a period close to 24 h, intensities of these metabolites, as obtained from their NMR spectra after normalizing and scaling all the spectra to make all 12 time points comparable, were plotted over the 12 time points recorded. Fig. 4 depicts (for the significant metabolites identified) the metabolite concentrations (as obtained from relative integrals) across 24 h, at 2 h intervals. As can be seen, all the metabolites show cycling with characteristic peaks and troughs during the 24 h. Time for peaks and troughs vary for individual metabolites. Kruskal–Wallis rank ANOVA was performed with time-point as the factor to confirm an effect of time of day followed by multiple comparisons to determine statistically significant peaks and troughs.| Metabolite | Peak (in ppm) used for quantification | Quantity present in | p-Value | BH corrected value | |
|---|---|---|---|---|---|
| ZT10 | ZT22 | ||||
| Sterol | 0.73–0.75 | 0.5404 ± 0.009 | 0.7706 ± 0.019 | 0.0012868 | 0.009651 |
| Fatty acid CH3 | 0.88–0.91 | 1.7923 ± 0.011 | 3.0091 ± 0.007 | 0.00066375 | 0.009651 |
| Fatty acid (CH2)n | 1.26–1.31 | 5.5942 ± 0.022 | 9.3585 ± 0.027 | 0.0021129 | 0.0105645 |
| Leucine | 0.94–0.96 | 5.7649 ± 0.016 | 6.3884 ± 0.027 | 0.017578 | 0.026367 |
| Valine | 0.97–0.99 | 4.2903 ± 0.008 | 4.9875 ± 0.110 | 0.036365 | 0.038824 |
| Isoleucine | 0.99–1.01 | 1.5361 ± 0.014 | 1.9324 ± 0.016 | 0.0071046 | 0.01522414 |
| Lactic acid | 1.32–1.34 | 8.5904 ± 0.019 | 6.2492 ± 0.025 | 0.0021129 | 0.01479325 |
| Alanine | 1.46–1.50 | 2.0623 ± 0.007 | 2.3564 ± 0.012 | 0.026435 | 0.03050192 |
| Lysine | 3.00–3.02 | 0.8342 ± 0.009 | 1.0170 ± 0.007 | 0.005627 | 0.01479325 |
| Creatine | 3.92–3.93 | 0.1574 ± 0.015 | 0.2724 ± 0.012 | 0.0097458 | 0.016243 |
| Glucose | 4.62–4.65 | 20.3064 ± 0.750 | 27.0216 ± 0.653 | 0.0059173 | 0.01479325 |
| Histidine | 7.09–7.11 | 0.7180 ± 0.012 | 0.6372 ± 0.021 | 0.024318 | 0.0303975 |
| Tryptophan | 7.30–7.32 | 0.4950 ± 0.006 | 0.3670 ± 0.012 | 0.021128 | 0.02881091 |
| NAD | 8.43–8.45 | 0.7381 ± 0.014 | 0.9793 ± 0.017 | 0.0094459 | 0.016243 |
| AMP | 8.59–8.60 | 1.3782 ± 0.011 | 1.5894 ± 0.009 | 0.038824 | 0.038824 |
We find that the concentrations of amino acids leucine, isoleucine, valine, lysine and alanine cycle in-phase, with higher levels during the night compared to day. This result provides confirmation of the method as one finds that molecules such as leucine, isoleucine and valine and all products of branched chain amino acid metabolism peak around the same time at night, http://www.genome.jp/keggbin/show_pathway?org_name=dme&mapno. While leucine, isoleucine and valine peaked around ZT18, which corresponds to mid-night, the other metabolites did not exhibit a sharp peak, but instead remained at high levels across a 6 h period. Such a peak near ZT18 was also detected for leucine, isoleucine and valine (peaks at ZT18.9, 18.8 and 18.6) by Minami and colleagues in mouse blood plasma26 while another study on human plasma also detected peak in leucine and lysine levels at ZT21.5 and ZT19 respectively29 suggesting a high degree of across-species conservation in the nature of oscillations for these metabolites. We also find that fatty acid levels build up gradually throughout the night and peaks late at night (ZT20–22). This is in agreement with a study which found that a large number of oscillating metabolites in the blood plasma of humans are fatty acids.30 Both histidine and tryptophan levels are higher during the day compared to night and in both cases no clear peak is seen. However, a previous study on mouse blood plasma suggests that tryptophan peaks around ZT19,26 differing quite dramatically from our results. Moreover, in other studies done on human plasma, concentrations of amino acids such as alanine and valine did not show a significant 24 h variation29,30 and tryptophan was not found to oscillate.29
For some other cycling metabolites, troughs appeared to be clearer than peaks. Around late evening (ZT10–14), glucose and AMP were found to be lowest, while creatine and sterol were lowest between ZT8–10 and ZT8–14 respectively. A recent study by Dallman and colleagues report that creatine levels in human saliva peaks at ZT16,30 while another study on mouse blood plasma reports a peak around ZT1526 which is in overall agreement with the profiles obtained from our samples, which also show creatine levels rising rapidly by that time. The study also indicates high levels of histidine in human saliva during the early part of the day similar to our own findings.30 Although many previous studies have suggested oscillations in glucose metabolism in mammals,47,48 to our knowledge, no previous study which used a metabolomics approach has reported rhythms in glucose levels. Our analysis of fly whole-body extracts suggests a clear trough in glucose levels at the end of day, in contrast to mice which show a trough in blood glucose levels late at night. However, since flies show crepuscular or mostly diurnal locomotor activity compared to mice, which are nocturnal, this difference in phase may be a reflection of differences in timing of glycogenolysis and gluconeogenesis pathways between these organisms. Our results show a steady rise in lactic acid levels through the day culminating in a peak shortly after lights-off (ZT14–16) with a trough beginning late at night (ZT22) and lasting until early half of the day (ZT4). Interestingly, our analysis picks up bimodality in the case of only one metabolite, NAD, which peaks both at mid day (ZT6) and also mid night (ZT18). NAD has previously been shown to oscillate with a circadian rhythm in mammalian cells (Nakahata et al. 2009, Science; Ramsey et al., 2009, Science) and is thought to be directly affected by nutritional status. Therefore, the peak at mid day may be attributed to feeding which is known to occur maximally immediately after dawn. However, the NAD peak at midnight is surprising and suggests that other processes that generate NAD occur at dusk.
Minami and colleagues showed that several hundred metabolites oscillate in the presence of LD cycles, of which a sizeable fraction (∼20%) continued to do so under constant dark conditions (DD) where no external time cues are available.26 The study proposed that the development of such high throughput methods to assay metabolite levels could, in future, be effective in detecting the state of the internal circadian clock in humans, diagnose clock disorders and prescribe personalised medication regimes. However, studies on humans and to a lesser extent mice are limited by the number of individuals that can be sampled and the kinds of experimental manipulations that can be performed. Hence our study using fruit flies with a high sample size (n = ∼40 per replicate per time point) and greater resolution provides an outline for future studies aimed at resolving the nature of the interaction between circadian clocks and metabolism. Researchers interested in the quantification of metabolite oscillations on a daily scale have relied on several methods to estimate metabolite levels in blood, saliva or other body tissues in humans24 and other model organisms or have relied on indirect evidence such as the oscillation in genes involved in metabolic processes.26,43 We propose that the use of high resolution NMR as described by our study will provide a much more accurate picture of oscillations of a large number of critical metabolites. As a proof-of-principle we present results from flies that have been subjected to a set of very strong time-cues namely light:dark and warm:cold cycles, which are known to synchronise not only behavioural rhythms such as locomotion and rest,49 eclosion50 and egg laying,51 but also rhythms of gene transcription that forms the basis of the core-circadian clock.52,53 Our results suggest that this method is robust and can reliably pick up oscillating metabolites from whole-body extracts of flies.
A previous study using the mass spectrometric approach has pointed out that the metabolome of adult male flies respond differentially when subjected to constant warmth (27 °C) as opposed to constant coolness at 18 °C.54 Specifically they identified purine nucleoside degradation pathway components adenosine, guanosine and adenine levels as being affected. Repeated exposures to cold temperatures are also known to modify energy reserves in flies,55 while in wasps Aphidius colemani, fluctuating temperatures were demonstrated to up-regulate metabolic pathways associated with glycolysis, Tricarboxylic acid (TCA) cycle and ATP synthesis.56 Interestingly our analysis did not reveal overlap with any of the above metabolites which may be due to the fact that temperature cycles that we employed are within the physiologically tolerable range as compared to many of the previous studies which examined stressful conditions of sustained warm or cold conditions or fluctuations where temperatures could rise or fall to stressful levels.
Interestingly several of the metabolites identified by our method overlap with those identified by previous studies which analyzed metabolomes from human and mice samples by mass spectrometry. Thus, it provides a starting point for future studies using genetic methods to modify genes or neurons important for the circadian clockwork or metabolic pathways or environmental regimes to disrupt rhythms in activity, sleep or feeding patterns and then apply this approach of NMR spectroscopy combined with multivariate statistical analysis to unravel their effects on the metabolome.
It is widely accepted that flies become mature adults around 5 days post-emergence, based on several features such as leveling off of sleep and activity counts at the age,5 sexual maturity of males,57 wing neurogenesis58 and hence we chose this age for our assays. Although age dependent changes have been reported in the metabolomic profile of flies, those changes have been documented over much larger ages spanning 3 days to 60 days.59 However, we cannot rule out the effect of small changes due to the difference in age across a 24 hour sampling period in the case of those metabolites that show a steady decline or increase across the 12 time points that we sampled.
Experimental methods
Flies from an outbred population of D. melanogaster (Meigen, 1830) maintained at a density of about 1200–1400 individuals per generation were used for this study. This population was formed by pooling flies from four previously described replicate laboratory populations maintained under 12:12 h LD cycles at 25 °C.60 The experimental population referred to as chrono control merged (CCM) continued to be maintained on a 21 day discrete generation cycle at 25 °C under 12:12 light:dark cycles. For the current experiment, eggs were sampled at the 29th generation since its creation. Eggs were collected on standard cornmeal agar medium at densities of approximately 50–60 per vial and development was completed in the same regime. Freshly emerged adult male flies were subjected to a five day regime where 12:12 h light:dark and warm (28 °C):cold (18 °C) cycles were superimposed such that the light period coincided with the high temperature duration and the dark period coincided with the low temperature duration. Five replicate vials containing 40 flies (6 day old) each were frozen at two h intervals starting at lights-ON. Flies were frozen at 12 consecutive time points during the course of 24 h. The frozen flies were then processed (Acetonitrile extraction) and lyophilized as described below.NMR sample preparation
The frozen flies from each sample were funneled into pre-labeled microfuge tubes containing 400 μl of ice-cold acetonitrile (50%) and homogenized using a battery-run homogenizer. Samples were then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C. The supernatant was transferred to another set of pre-labeled and pre-weighed microfuge tubes, and stored at −80 °C until lyophilized. The lyophilized samples were then used for NMR analysis. Prior to NMR measurements, the samples were rehydrated in 500 μl of 50 mM phosphate buffer made up in D2O (pH 7.4), and 450 μl of the sample was transferred to a 5 mm NMR tube. 1 mg ml−1 of 3-(trimethylsilyl)-propionic acid-D4, sodium salt (TMSP) was added as a chemical shift reference.
000 rpm for 10 min at 4 °C. The supernatant was transferred to another set of pre-labeled and pre-weighed microfuge tubes, and stored at −80 °C until lyophilized. The lyophilized samples were then used for NMR analysis. Prior to NMR measurements, the samples were rehydrated in 500 μl of 50 mM phosphate buffer made up in D2O (pH 7.4), and 450 μl of the sample was transferred to a 5 mm NMR tube. 1 mg ml−1 of 3-(trimethylsilyl)-propionic acid-D4, sodium salt (TMSP) was added as a chemical shift reference.
1D and 2D NMR spectroscopy
NMR spectra were recorded at 298 K on a Bruker Biospin 600 MHz Avance-III spectrometer operating at a 1H NMR frequency of 600.13 MHz, equipped with a 5 mm TXI probe. Gradient shimming was performed prior to signal acquisition. D2O was used to provide an internal lock. 1D 1H NMR spectra were acquired using a water suppressed Car–Purcell–Meiboom–Gill (CPMG) spin-echo pulse sequence optimized with a spin-echo delay t of 300 μs and n = 400 and a total spin–spin relaxation delay (2nt) time of 240 ms to achieve attenuation of fast-relaxing broad signals from larger molecules. Water suppression of the residual water signal was achieved with a pre-saturation sequence with low-power selective irradiation at 4.7 ppm during the recycle delay. The proton spectra were collected with a 90° pulse width of 7.4 μs, a recycle delay of 2 s, 16 scans, 16 K data points and a spectral width of 12 ppm. Data were zero-filled by a factor of 2 and the FIDs were multiplied by an exponential weighting function equivalent to a line broadening of 1 Hz prior to Fourier transformation. The spectra were phase and baseline corrected and referenced to the TMSP resonance at 0.00 ppm.For resonance assignment and metabolite identification, two-dimensional NMR spectra were recorded, including 1H–1H correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), and 1H–13C heteronuclear coherence spectroscopy (HSQC, HMQC). 2D 1H–13C heteronuclear spectra were obtained with a spectral width of 12 ppm and 200 ppm in the proton and carbon dimensions respectively, 1 K data points, 16 scans, 256t−1 increments and a recycle delay of 2 s. The COSY and TOCSY spectra were acquired with a spectral width of 12 ppm in both dimensions, 1 K data points, 16 scans and 256t−1 increments.
Statistical analysis
1D 1H NMR spectra were used for statistical analysis. The variation and possible cycling in concentrations of metabolites were observed by sampling the flies after every two hours over a 24 hour LD warm:cold cycle so as to obtain fly samples at twelve different time points (ZT0 to ZT22).The full NMR spectra, without binning, were converted into the ASCII format and imported into MATLAB for alignment using the icoshift algorithm.46 Prior to alignment, spectral regions between 4.66 and 4.8 ppm were excluded from the analysis, to mitigate errors due to any residual peak from the suppressed water signal. Regions between 0.6 ppm and 9.5 ppm were used for analysis after removing high and low-field regions where no signal (except the TMSP reference peak) was present. Data were normalized to the total area to compensate for possible differences in signal-to-noise ratios between spectra and to prevent separation due to variations in the amounts of sample.
Multivariate statistical analysis was performed on the resulting data using the SIMCA 14.0 software (Umetrics, Umea, Sweden). The data were first imported into SIMCA, Pareto scaled and analyzed using the unsupervised pattern recognition method of principal component analysis (PCA). The PCA was used to compare the samples from all twelve time points and also helped to remove outliers, defined in the data as observations located outside the 95% confidence region of the Hotellings T2 ellipses in the PCA score plots. Such outliers were excluded from further analysis. The output from PCA analysis consists of scores plots and loadings plots, providing information about grouping in the data due to metabolic similarities and identifying variables which are responsible for patterns obtained in the scores plots, respectively. The performance of the PCA model was evaluated using the correlation coefficient R2 and using the method of 7-fold internal cross-validation of SIMCA for obtaining the coefficient Q2. R2 defines the variance explained, whereas Q2 defines the variance predicted by the model. A good prediction model is expected when Q2 > 0.5, and with Q2 > 0.9, the model is considered to have an excellent predictive ability.
PCA was followed by the supervised pattern recognition method of orthogonal projections to latent structure-discriminant analysis (OPLS DA), which maximizes the class discrimination. OPLS-DA analysis was performed to find out how different were the two 12 h apart time points (ZT10 and ZT22) and to identify metabolites that were responsible for the separations. Such metabolites can be considered to change their concentrations during the 24 h when flies are exposed to a 12:12 h LD warm:cold cycle. Similar to the PCA score plot, the OPLS-DA scores plot gives Hotelling's T2 regions, shown as an ellipse in the scores plot, defining the 95% confidence interval of the modelled variation. The loadings plot was used to identify variables (metabolites) responsible for the maximum separation of time points. Significant metabolites were ranked according to their variable influence on the projection (VIP) score. VIP analysis displays the metabolites ordered according to their influence on group separation, with metabolites arranged according to their VIP values, and the y-axis denoting their relative intensities. VIP (variable importance projection) of the first principal component in the OPLS-DA model (VIP > 1.0) is used to screen significantly different metabolites. The quality of the model was described by R2X and Q2 values, defining the variance explained (indicating goodness of fit) and variance predicted by the model (predictability) respectively. The significance test of the model was performed using CV-ANOVA (cross-validated ANOVA)61 in the SIMCA software, where a p-value <0.05 was considered to be statistically significant and clearly validates the OPLS-DA model. Permutation analysis was further performed on the best model using 1000 permutation tests with a threshold p value of <0.05 indicating that none of the results are better than the original one.62,63
Hierarchical cluster analysis (HCA) was performed to create a dendrogram, using the complete linkage algorithm and the Euclidean distance between OPLS-DA scores for each replicate of the selected time points. For better visualization, a fan (polar) dendrogram was also generated to compare all time points and to identify metabolite cycling at different time points using a MATLAB script (http://www.mathworks.com/matlabcentral/fileexchange/21983-draw-a-polar-dendrogram).
Univariate analysis was performed to check for statistical significance of the differences in the metabolite levels. For comparison of all the time points together, ANOVA (p < 0.01) was used, which can determine whether the comparison between multi-group analysis is significant or not. This was followed by post hoc analysis in order to perform all pairwise comparisons between group means by a least significant difference (LSD) t-test. For comparison involving two time points in OPLS-DA, the t-test was performed on the identified metabolites (from the integral area of 1H NMR peaks corresponding to each metabolite). Metabolites having VIP > 1 and t-test p value < 0.05 were selected. Since the t-test is performed separately on each metabolite independent of other metabolites, the chance of false positives is proportional to the number of tests performed. Multiple testing correction was hence applied, which adjusts the individual p value for each metabolite to keep the overall error rate (or false positive rate) to be less than or equal to the user-specified p value cut-off. Thus to cross-check that all the identified metabolites were statistically significant, a multiple hypothesis test correction using the method of Benjamini–Hochberg was also performed with a value of 0.05 selected as the level of significance.64
Metabolite databases & software
Metabolite fingerprinting for the Drosophila NMR spectra was done by checking identified metabolite peaks with standard NMR metabolite data deposited in databases such as the Biological Magnetic Resonance Data Bank (BMRB), http://www.bmrb.wise.edu and the Madison Metabolomics Consortium Database (MMCD), http://mmcd.nmrfam.wise.edu.Conclusions
Our study demonstrates diurnal rhythms in concentrations of several metabolites over a 24 h time period in D. melanogaster, using NMR-based metabolomics in combination with multivariate analysis. This study paves the way for use of NMR-based metabolomics in examining questions related to the circadian clock and metabolome level changes, which has previously been studied using less accurate methods. While it would not be appropriate at this juncture to compare across species without proper validation using multiple approaches, it is nevertheless heartening that several oscillating metabolites identified using our method have also been identified by LC-MS or GC-MS methods in mice and human tissues. This study provides impetus to carry out future investigations where circadian clock genes or circuits can be modified in a tissue or a time dependent manner following which one can profile changes in critical metabolites. Furthermore, we can target specific metabolic pathways by genetic manipulation or introduce specific feeding/fasting or exercise/stress regimens which could be expected to modify the metabolic profile of organisms. This experimental organism and approach could also be extended to include pharmacological intervention and screen for small molecules that can provide effective modulation of metabolic pathways. Hence, we believe this to be the first step in the direction of exciting future possibilities.Acknowledgements
The experiments were performed on a Bruker Avance-III 600 MHz FT-NMR spectrometer at the NMR Research Facility at IISER Mohali. We thank Dr Ranga Udaykumar, for use of VirTis BenchTop Lyophilizer and Dr Ravi Manjithaya for use of a −80 refrigerator. We thank Joydeep De for help with initial experiments and Sheetal Potdar for help during the assay and suggestions on the manuscript. We thank Harpreet Singh for help with implementing Matlab scripts to make the polar dendrogram.References
- G. Asher and U. Schibler, Cell Metab., 2011, 13, 125–137 CrossRef CAS PubMed.
- U. Albrecht, Neuron, 2012, 74, 246–260 CrossRef CAS PubMed.
- A. Eskin, Fed. Proc., 1979, 38, 2570–2572 CAS.
- S. Sahar and P. Sassone-Corsi, Trends Endocrinol. Metab., 2012, 23, 1–8 CrossRef CAS PubMed.
- K. A. Lamia, U. M. Sachdeva, L. DiTacchio, E. C. Williams, J. G. Alvarez, D. F. Egan, D. S. Vasquez, H. Juguilon, S. Panda, R. J. Shaw, C. B. Thompson and R. M. Evans, Science, 2009, 326, 437–440 CrossRef CAS PubMed.
- C. Canto, Z. Gerhart-Hines, J. N. Feige, M. Lagouge, L. Noriega, J. C. Milne, P. J. Elliott, P. Puigserver and J. Auwerx, Nature, 2009, 458, 1056–1060 CrossRef CAS PubMed.
- B. Marcheva, K. M. Ramsey, E. D. Buhr, Y. Kobayashi, H. Su, C. H. Ko, G. Ivanova, C. Omura, S. Mo, M. H. Vitaterna, J. P. Lopez, L. H. Philipson, C. A. Bradfield, S. D. Crosby, L. JeBailey, X. Wang, J. S. Takahashi and J. Bass, Nature, 2010, 466, 627–631 CrossRef CAS PubMed.
- F. W. Turek, C. Joshu, A. Kohsaka, E. Lin, G. Ivanova, E. McDearmon, A. Laposky, S. Losee-Olson, A. Easton, D. R. Jensen, R. H. Eckel, J. S. Takahashi and J. Bass, Science, 2005, 308, 1043–1045 CrossRef CAS PubMed.
- M. F. Ceriani, J. B. Hogenesch, M. Yanovsky, S. Panda, M. Straume and S. A. Kay, J. Neurosci., 2002, 22, 9305–9319 CAS.
- N. Cermakian, L. Monaco, M. P. Pando, A. Dierich and P. Sassoni-Corsi, EMBO J., 2001, 20, 3967–3974 CrossRef CAS PubMed.
- S. Panda, M. P. Antoch, B. H. Miller, A. I. Su, A. B. Schook, M. Straume, P. G. Schultz, S. A. Kay, J. S. Takahashi and J. B. Hogenesch, Cell, 2002, 109, 307–320 CrossRef CAS.
- K. Xu, J. R. DiAngelo, M. E. Hughes, J. B. Hogenesch and A. Sehgal, Cell Metab., 2011, 13, 639–654 CrossRef CAS PubMed.
- K. J. Venken, J. H. Simpson and H. J. Bellen, Neuron, 2011, 72, 202–230 CrossRef CAS PubMed.
- K. Xu, X. Zheng and A. Sehgal, Cell Metab., 2008, 8, 289–300 CrossRef CAS PubMed.
- K. L. Eckel-Mahan, V. R. Patel, S. de Mateo, R. Orozco-Solis, N. J. Ceglia, S. Sahar, S. A. Dilag-Penilla, K. A. Dyar, P. Baldi and P. Sassone-Corsi, Cell, 2013, 155, 1464–1478 CrossRef CAS PubMed.
- Y. Nakahata, M. Kaluzova, B. Grimaldi, S. Sahar, J. Hirayama, D. Chen, L. P. Guarente and P. Sassone-Corsi, Cell, 2008, 134, 329–340 CrossRef CAS PubMed.
- J. S. O'Neill, E. S. Maywood, J. E. Chesham, J. S. Takahashi and M. H. Hastings, Science, 2008, 320, 949–953 CrossRef PubMed.
- K. M. Ramsey, J. Yoshino, C. S. Brace, D. Abrassart, Y. Kobayashi, B. Marcheva, H. K. Hong, J. L. Chong, E. D. Buhr, C. Lee, J. S. Takahashi, S. Imai and J. Bass, Science, 2009, 324, 651–654 CrossRef CAS PubMed.
- A. B. Reddy, N. A. Karp, E. S. Maywood, E. A. Sage, M. Deery, J. S. O'Neill, G. K. Wong, J. Chesham, M. Odell, K. S. Lilley, C. P. Kyriacou and M. H. Hastings, Curr. Biol., 2006, 16, 1107–1115 CrossRef CAS PubMed.
- A. N. Dodd, M. J. Gardner, C. T. Hotta, K. E. Hubbard, N. Dalchau, J. Love, J. M. Assie, F. C. Robertson, M. K. Jakobsen, J. Goncalves, D. Sanders and A. A. Webb, Science, 2007, 318, 1789–1792 CrossRef CAS.
- Y. Nakahata, S. Sahar, G. Astarita, M. Kaluzova and P. Sassone-Corsi, Science, 2009, 324, 654–657 CrossRef CAS PubMed.
- S. Imai, C. M. Armstrong, M. Kaeberlein and L. Guarente, Nature, 2000, 403, 795–800 CrossRef CAS PubMed.
- G. Asher, D. Gatfield, M. Stratmann, H. Reinke, C. Dibner, F. Kreppel, R. Mostoslavsky, F. W. Alt and U. Schibler, Cell, 2008, 134, 317–328 CrossRef CAS PubMed.
- H. K. Kim, Y. H. Choi and R. Verpoorte, Nat. Protoc., 2010, 5, 536 CrossRef CAS PubMed.
- O. Beckonert, H. C. Keun, T. M. D. Ebbel, J. Bundy, E. Holmes, J. C. Lindon and J. K. Nicholson, Nat. Protoc., 2007, 2, 2692–2703 CrossRef CAS PubMed.
- Y. Minami, T. Kasukawa, Y. Kakazu, M. Iigo, M. Sugimoto, S. Ikeda, A. Yasui, G. T. van der Horst, T. Soga and H. R. Ueda, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 9890–9895 CrossRef CAS PubMed.
- M. Hatori, C. Vollmers, A. Zarrinpar, L. DiTacchio, E. A. Bushong, S. Gill, M. Leblanc, A. Chaix, M. Joens, J. A. Fitzpatrick, M. H. Ellisman and S. Panda, Cell Metab., 2012, 15, 848–860 CrossRef CAS PubMed.
- K. L. Eckel-Mahan, V. R. Patel, R. P. Mohney, K. S. Vignola, P. Baldi and P. Sassone-Corsi, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 5541–5546 CrossRef CAS PubMed.
- J. E. Ang, V. Revell, A. Mann, S. Mantele, D. T. Otway, J. D. Johnston, A. E. Thumser, D. J. Skene and F. Raynaud, Chronobiol. Int., 2012, 29, 868–881 CrossRef CAS PubMed.
- R. Dallmann, A. U. Viola, L. Tarokh, C. Cajochen and S. A. Brown, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2625–2629 CrossRef CAS PubMed.
- T. Kasukawa, M. Sugimoto, A. Hida, Y. Minami, M. Mori, S. Honma, K. Honma, K. Mishima, T. Soga and H. R. Ueda, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 15036–15041 CrossRef CAS PubMed.
- A. Malmendal, J. Overgaard, J. G. Bundy, J. G. Sorensen, N. C. Nielsen, V. Loeschcke and M. Holmstrup, Am. J. Physiol.: Regul., Integr. Comp. Physiol., 2006, 291, R205–R212 CrossRef CAS PubMed.
- J. Overgaard, A. Malmendal, J. G. Sorensen, J. G. Bundy, V. Loeschcke, N. C. Nielsen and M. Holmstrup, J. Insect Physiol., 2007, 53, 1218–1232 CrossRef CAS PubMed.
- K. S. Pedersen, T. N. Kristensen, V. Loeschcke, B. O. Petersen, J. O. Duus, N. C. Nielsen and A. Malmendal, Genetics, 2008, 180, 1233–1243 CrossRef CAS PubMed.
- J. D. Feala, L. Coquin, D. Zhou, G. G. Haddad, G. Paternostro and A. D. McCulloch, BMC Syst. Biol., 2009, 3, 91 CrossRef PubMed.
- J. D. Feala, J. H. Omens, G. Paternostro and A. D. McCulloch, Ann. N. Y. Acad. Sci., 2008, 1123, 169–177 CrossRef CAS PubMed.
- J. D. Feala, L. Coquin, A. D. McCulloch and G. Paternostro, Mol. Syst. Biol., 2007, 3, 99 CrossRef PubMed.
- A. Malmendal, J. G. Sorensen, J. Overgaard, M. Holmstrup, N. C. Nielsen and V. Loeschcke, Naturwissenschaften, 2013, 100, 417–427 CrossRef CAS PubMed.
- L. Coquin, J. D. Feala, A. D McCulloch and G. Paternostro, Mol. Syst. Biol., 2008, 4, 1–13 CrossRef PubMed.
- P. Sarup, S. M. M. Pedersen, N. C. Nielsen, A. Malmendal and V. Loeschcke, PLoS One, 2012, 7, 1–11 Search PubMed.
- A. C. Keene, E. R. Duboue, D. M. McDonald, M. Dus, G. S. Suh, S. Waddell and J. Blau, Curr. Biol., 2010, 20, 1209–1215 CrossRef CAS PubMed.
- J. R. DiAngelo, R. Erion, A. Crocker and A. Sehgal, PLoS One, 2011, 6, e19921 CAS.
- D. J. Seay and C. S. Thummel, J. Biol. Rhythms, 2011, 26, 497–506 CrossRef PubMed.
- T. Yoshii, S. VAnin, R. Costa and C. Helfrich-Förster, J. Biol. Rhythms, 2009, 24, 452–464 CrossRef PubMed.
- O. Cloarec, M. E. Dumas, J. Trygg, A. Craig, R. H. Barton, J. C. Lindon, J. K. Nicholson and E. Holmes, Anal. Chem., 2005, 77, 517–552 CrossRef CAS PubMed.
- F. Savorani, G. Tomasi and S. B. Engelsen, J. Magn. Reson., 2010, 202, 190–202 CrossRef CAS PubMed.
- R. Feneberg and B. Lemmer, Chronobiol. Int., 2004, 21, 521–538 CrossRef CAS PubMed.
- A. Kalsbeek, S. La Fleur, C. Van Heijningen and R. M. Buijs, J. Neurosci., 2004, 24, 7604–7613 CrossRef CAS PubMed.
- T. Yoshii, C. Hermann and C. Helfrich-Förster, J. Biol. Rhythms, 2010, 25, 387–398 CrossRef PubMed.
- K. L. Nikhil, G. Goirik, K. Ratna and V. K. Sharma, J. Biol. Rhythms, 2014, 29, 427–441 CrossRef CAS PubMed.
- N. N. Kannan, R. Reveendran, S. Hari Dass, T. Manjunatha and V. K. Sharma, J. Insect Physiol., 2012, 58, 245–255 CrossRef CAS PubMed.
- F. T. Glaser and R. Stanewsky, Curr. Biol., 2005, 15, 1352–1363 CrossRef CAS PubMed.
- H. Sehadova, F. T. Glaser, C. Gentile, A. Simoni, A. Giesecke, J. T. Albert and R. Stanewsky, Neuron, 2009, 64, 251–266 CrossRef CAS PubMed.
- R. Hariharan, J. M. Hoffman, A. S. Thomas, Q. A. Soltow, D. P. Jones and D. E. L. Promislow, BMC Syst. Biol., 2014, 8, 139 CrossRef PubMed.
- K. E. Marshall and B. J. Sinclair, Proc. R. Soc. London, Ser. B, 2010, 277, 963–969 CrossRef PubMed.
- H. Coline, T. T. An Nguyen, C. Cloutier, D. Michaud and T. Hance, Mol. Biol., 2007, 37, 1177–1188 Search PubMed.
- N. Svetec and J. F. Ferveur, J. Exp. Biol., 2005, 5, 891–898 CrossRef PubMed.
- S. Ben Rokia-Mille, S. Tinette, G. Engler, L. Arthaud, S. Tares and A. Robichon, PLoS One, 2008, 3, e2395 Search PubMed.
- J. M. Hoffman, Q. A. Soltow, Sh. Li, A. Sidik, D. P. Jones and D. E. L. Promislow, Aging Cell, 2014, 13, 596–604 CrossRef CAS PubMed.
- N. N. Kannan, K. M. Vaze and V. K. Sharma, J. Exp. Biol., 2012, 215, 3527–3534 CrossRef PubMed.
- L. Eriksson, J. Trygg and S. Wold, J. Chemom., 2008, 22, 594–600 CrossRef CAS PubMed.
- R. G. Brereton, Chemometrics: Data analysis for the laboratory and chemical plant, John Wiley and Sons, Chichester, England, 2003 Search PubMed.
- J. Shao, J. Am. Stat. Assoc., 1993, 88, 486–494 CrossRef PubMed.
- Y. Benjamini and Y. Hochberg, J R Stat Soc Series B, 1995, 57, 289–300 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1 shows the 1D 1H NMR spectra of D. melanogaster recorded at 600 MHz. Table S1 lists metabolites identified from 1D and 2D NMR experiments on D. melanogaster. Table S2 lists metabolites identified to be responsible for separation of all twelve time points as obtained from ANOVA (p < 0.01). Fig. S3–S6 contain OPLS-DA scores plot for comparison between ZT8–ZT20, ZT6–ZT18, ZT4–ZT16, ZT0–ZT22 and ZT2–ZT22 time points respectively. Table S3 lists metabolites identified from the t-test (p < 0.01) to be responsible for separation between ZT6–ZT18, ZT4–ZT16, ZT0–ZT22 and ZT2–ZT22 time points. See DOI: 10.1039/c5mb00386e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |