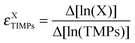Open Access Article
Open Access ArticleS100A4 and its role in metastasis – simulations of knockout and amplification of epithelial growth factor receptor and matrix metalloproteinases†
Antoine
Buetti-Dinh
*abc,
Igor V.
Pivkin
bd and
Ran
Friedman
*ac
aDepartment of Chemistry and Biomedical Sciences, Linnæus University, Kalmar, Sweden. E-mail: antoine.buetti@lnu.se; ran.friedman@lnu.se
bInstitute of Computational Science, Faculty of Informatics, Università della Svizzera Italiana, Lugano, Switzerland
cCentre of Excellence for Biomaterials Chemistry, Linnæus University, Kalmar, Sweden
dSwiss Institute of Bioinformatics, Lausanne, Switzerland
First published on 3rd June 2015
Abstract
The calcium-binding signalling protein S100A4 enhances metastasis in a variety of cancers. Despite a wealth of data available, the molecular mechanism by which S100A4 drives metastasis is unknown. Integration of the current knowledge defies straightforward intuitive interpretation and requires computer-aided approaches to represent the complexity emerging from cross-regulating species. Here we carried out a systematic sensitivity analysis of the S100A4 signalling network in order to identify key control parameters for efficient therapeutic intervention. Our approach only requires limited details of the molecular interactions and permits a straightforward integration of the available experimental information. By integrating the available knowledge, we investigated the effects of combined inhibition of signalling pathways. Through selective knockout or inhibition of the network components, we show that the interaction between epidermal growth factor receptor (EGFR) and S100A4 modulates the sensitivity of angiogenesis development to matrix metalloproteinases (MMPs) activity. We also show that, in cells that express high EGFR, MMP inhibitors are not expected to be useful in tumours if high activity of S100A4 is present.
1 Introduction
S100A4 belongs to the S100 family of low-molecular weight calcium-binding proteins that transmit cellular signals through conformational changes mediated by Ca2+ and other ions.1 There are more than 20 known S100 proteins in humans, many of which are tissue- or cell-type specific and have altered expression in some types of cancer.2–4 S100A4 has been reported to be involved in several different processes related to cancer progression.5 In cancer tissues, S100A4 has been found in cytoplasm, nucleus and also in the extracellular matrix.5 The protein is expressed by a variety of cell types in the tumour microenvironment of human breast cancer6 and its increased expression is associated with human colorectal adenocarcinomas.7,8 Early studies have shown that increased levels of S100A4 induce a metastatic phenotype in rodent models of mammary carcinogenesis.9,10 Moreover, knockdown of S100A4 suppresses cell migration and metastasis in osteosarcoma cell lines11 and reduces cell growth and motility in human pancreatic cancer cells.12 S100A4 is preferentially expressed in cells with motile phenotype5 and it influences motility13–15 and invasion16–18via interactions with myosin IIA (myosin-9) and MMPs, respectively. Several studies report that S100A4 promotes angiogenesis.19,20 Tumours with elevated levels of S100A4 show high vessel density in breast cancer21 where S100A4 stimulates angiogenesis via interactions with annexin II20 and osteopontin (OPN).22 MMPs and their natural inhibitors, tissue inhibitors of matrix metalloproteinases (TIMPs), play a major role in these processes by regulating the degradation of the extracellular matrix and consequently facilitating (or preventing) invasion and tissue remodelling. Interactions between S100A4, MMPs and epithelial–mesenchymal transition (EMT) target genes such as β-catenin and E-cadherin have been shown in different systems.4,5,23 It has also been verified in several models that S100A4 knockdown reduces the expression of MMP genes, thereby suppressing cell migration.18,24 Extracellular S100A4 appears to be involved in EGFR signalling by interacting with EGFR/ErbB2 ligands.25 These interactions have been shown to enhance S100A4 expression26 suggesting potential regulation through positive feedback.4 The possibility of interactions between p53 and S100A4 has previously been debated27,28 and is supported by recent findings showing that S100A4 interacts with p53 and MDM2, indicating that S100A4 promotes p53 degradation.29Despite abundant data on S100A4 and its interacting partners, the regulatory aspects underlying the bridging role that S100A4 represents between metastasis and angiogenesis is still poorly understood. Consequently, the development of therapies aiming to block targeted components of the S100A4 network is hampered by insufficient understanding of its complex compensatory signalling. Here we apply network analysis to predict the effect of (combined) targeted inhibition in silico and to identify the mechanisms and network's control points that are relevant for the development of successful therapeutic strategies.
2 Methods
2.1 S100A4 network
A signalling network model for S100A4 based on the experimental evidence mentioned previously together with a set of derived networks corresponding to the different biological situations described in the “Results” section were used to study the effects of gene knockout and overexpression on the S100A4 network and to investigate potential therapeutic strategies by selective inhibition of some of the network's components.2.2 Modelling framework
We used a quantitative phenomenological modelling framework composed of modules for numerical simulation and analysis to study biological networks in a flexible way (details are found in the companion article30). The method allows to efficiently carry out sensitivity analysis of biological networks and thereby to identify key control points where the effect of addition, removal and inhibition of components that can have a high impact on the endpoint(s). Modifications can easily be applied to pre-existing settings, and new entries can be rather easily integrated in the model. Consequently, the effect of gene knockout, overexpression and other perturbations can be tested without a detailed mechanistic knowledge of the underlying interactions. Parameter ranges can be adapted according to available knowledge: from several orders of magnitude in case of poorly characterized processes, to a much narrower range if suggested by accurate experimental information. The system's response to parameter variation reveals the role of different components. The results are summarized through graphical representations (principal component analysis (PCA) and sensitivity landscape plots). Through this procedure, we can identify control points in the network such as switches (sensitive regions) where small changes yield large effects on the biological outcome (e.g., cell dissociation) or buffers (paths that are robust with respect to external perturbations).3 Results
We carried out simulations of the S100A4 network as presented in Fig. 1. Thereby, we investigated the combined effects of gene knockout and targeted inhibition by removing nodes and constraining the activities of certain components of the network. Simulation of such modified systems enables the characterization of the effects of therapeutic interventions in quantitative terms and the identification of potential resistance mechanisms. In the following sections we detail the in silico response of a series of cases where the network model of S100A4 (see scheme in Fig. 1) is modified by removal or alteration of the most relevant network components (S100A4, MMPs, EGFR, NF-κB).3.1 Simulations with a larger network
The S100A4 network scheme represented in Fig. 1 is an approximation of the main processes influencing cell dissociation and capillary growth. To ensure that the outcome of our simulations is independent of the set up of the network, we tested our approach with an extended S100A4 network (see Fig. ESI 1 (ESI†)). The extended network has been enlarged according to references31–38 and includes 9 additional nodes and 13 additional reactions compared to the network represented in Fig. 1 (this corresponds to an increase of 60% and 42% for nodes and reactions, respectively). We note that in robustness tests, a network able to tolerate a variation of 5–20% in the number of the nodes is considered highly robust.39,40 The results obtained with the enlarged network (see Fig. ESI 2 (ESI†)) are consistent with the ones that correspond to the scheme of Fig. 1 (compare Fig. ESI 2 (ESI†) with Fig. 2 and 3 of the companion article30). This indicates that the effects of S100A4 on the network are reproducible over a wide range of network components considered in the network.3.2 S100A4 knockout
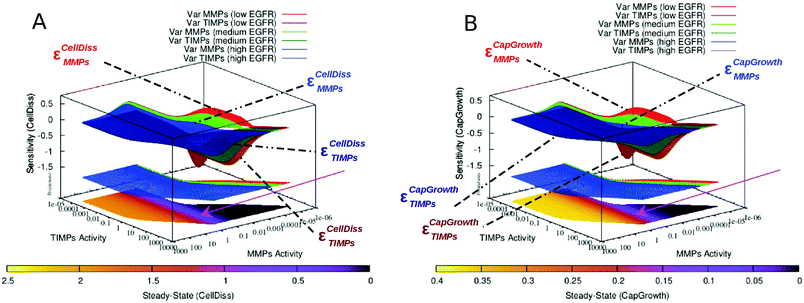
We emulated S100A4 gene knockout by removing S100A4 (intra- and extracellular) from the node list and consequently all in- and out-pointing edges. This way, we mimic cancer cells that are devoid of S100A4 activity or treatment by an effective S100A4 inhibitor, which may become available in the future.41,42 We then ran simulations with different levels of EGFR activity. Compared to simulations of the complete network set with an initially low S100A4 basal activity level, in response to increasing EGFR activity, the sensitivity to MMPs and TIMPs increases moderately for cell dissociation and strongly for capillary growth whereas their corresponding steady-state levels are not significantly affected (see Fig. 2). We have previously reported that, in the presence of S100A4, the sensitivity of capillary growth to MMPs and TIMPs showed a complex pattern, where two stable regions (insensitive to MMPs and TIMPs activities) could be observed. Knocking out S100A4 abolished this pattern. Instead, a general decrease in sensitivity to MMPs and TIMPs was observed. Interestingly, both MMPs and TIMPs can adopt broader range of activities when S100A4 is knocked down, whereas only the activity of MMPs is shifted to higher values in response to increased levels of EGFR (see the projections on the plane corresponding to variable EGFR activity in Fig. 2).3.3 Inhibition of MMPs
MMPs are released by the tumour microenvironment and play an important role in cancer progression by enhancing cell motility and invasion.6,43,44 Inhibition of metalloproteinases has been the focus of diverse therapeutic strategies against cancer.45 We therefore simulated MMPs inhibition by eliminating from the full network model the links that influence its activity and constraining its steady-state level to a constant low value of 0.0001. We then followed on the variation of S100A4 in combination with increasing EGFR (see section “S100A4 knockout”). TIMPs do not influence the system (see Fig. ESI 3 (ESI†)). In the absence of MMPs, S100A4 activity levels correlate to cell dissociation and capillary growth. MMPs inhibition increases the sensitivity of capillary growth to variations, whereas that of cell dissociation remains unaltered. In addition, a sensitivity barrier (i.e., a peak of the calculated sensitivity function) between low and high steady-state values of both variables CellDiss and CapGrowth can be observed. The barrier decreases proportionally to EGFR activity (see Fig. 3).3.4 S100A4 knockout and inhibition of MMPs
We subsequently combined the two approaches described in sections “S100A4 knockout” and “Inhibition of MMPs” by simulating systematic variation of EGFR while modulating the activity of NF-κB (TIMPs do not influence the system as in section “Inhibition of MMPs”, see Fig. ESI 4 (ESI†)). This combined approach appears to successfully counter both cell dissociation and capillary growth as indicated by lower steady state levels (see Fig. ESI 4 (ESI†)) and an increase in the sensitivity as a function of EGFR activity (see Fig. 4). The latter is an indication that the system can be under control. However, the system is still able to sustain a regime dictated by high levels of cell dissociation and capillary growth through (very) high activity of EGFR (Fig. ESI 4 (ESI†)). It is not clear if such high values of EGFR can be maintained in cells. Thus, it may be possible to limit or postpone metastasis by MMP inhibitors in the absence of S100A4. A sensitivity barrier between low and high steady-state values of CellDiss and CapGrowth variables is inversely proportional to the activity of NF-κB.3.5 Inhibition of EGFR-mediated feedback of S100A4
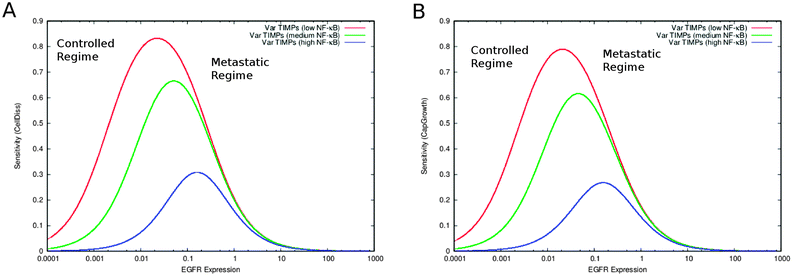
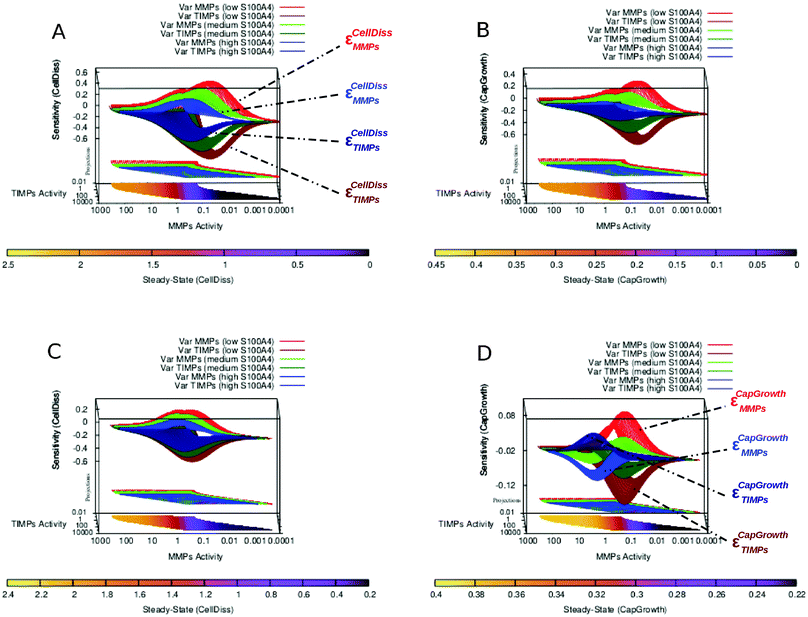
EGFR-mediated feedback of S100A4 has previously been suggested.4,25,26 We consequently simulated the system without the interactions between EGFR and S100A4 (both intra- and extracellular). Inhibiting EGFR increases the sensitivity of cell dissociation and capillary growth consequently improving controllability. At the same time it restricts MMPs and TIMPs steady-state to higher ranges (Fig. 5A and B compared to Fig. 5C and D). With and without the EGFR-mediated feedback of S100A4, an increase in the concentration of biologically active S100A4 causes a homogeneous reduction of sensitivity without affecting the corresponding steady-state values. In addition, EGFR inhibition was sufficient to abolish the formation of multiple regions sensitive to capillary growth separated by the near-zero sensitivity boundaries as observed in Fig. 5D, highlighting the role of the EGFR-mediated positive feedback in the co-regulation between S100A4 and EGFR reported previously4 and observed in the PCA applied to steady-state and sensitivity data as a group of variables including S100A4 together with EGFR and NF-κB closely linked to the variable representing capillary growth (Fig. ESI 8 and Text ESI 1.5 (ESI†)).3.6 The pattern of multiple sensitivity regions depends on EGFR activity
We further investigated in more details the phenomenon described in the previous section. Besides increasing sensitivity of cell dissociation and restricting MMPs and TIMPs to higher steady-state ranges, a more complex effect is observed in the alteration of the capillary growth sensitivity landscape. While no relevant change in CellDiss and CapGrowth steady-state values is observed, the multistable character of the system presenting multiple sensitive regions that rearrange as a function of S100A4 activity (see Fig. 5D) disappears, yielding a homogeneous surface over a wider sensitivity range (Fig. 5B). A more detailed representation of EGFR-mediated feedback as applied to the sensitivity of capillary growth is shown in Fig. 6. Multistable equilibria are observed at intermediate activity of S100A4 (Fig. 6A). This branched pattern of states characterised by low sensitivity disappears with inhibition of EGFR-mediated feedback (Fig. 6B) and consistent behaviour is observable in the PCA.4 Discussion
S100A4 is used as a prognostic marker for metastatic tumours. This small, metastasis promoting protein occupies a central place in a dense interaction network of cancer-related processes. Extensive experimental evidence connects S100A4 to angiogenesis, invasion and enhanced cell motility in a variety of cancers. In spite of the abundant data, integration of current knowledge in a therapeutic context presents a challenge. Several S100A4-interacting partners have been used as targets for therapeutic intervention in the past, but the intricacy of such signalling pathways often leads to drug resistance. Quantitative insights are therefore necessary to understand dosage and compensation effects that emerge from complex signalling interactions. Using network-based simulations and biologically relevant yet simplifying assumptions, we could investigate the combined effects of inhibition without detailed knowledge of the interactions between the nodes. Nonlinear effects resulting from feedbacks, pleiotropy and redundancy could therefore be taken into consideration in a global context.4.1 EGFR-mediated Feedback of S100A4 is responsible for multistable buffering patterns
The knockout of S100A4 or the inhibition of the EGFR-mediated feedback of S100A4 causes the loss of the multistable buffering pattern previously observed in the full S100A4 network (Fig. 5 and 6). We showed that the positive feedback plays a crucial role in the co-regulation between S100A4 and EGFR, reported earlier.25,26 This behaviour was further investigated by focussing on a near-zero sensitivity section at intermediate activity of S100A4 (Fig. 5). A branched, broad buffering area insensitive to MMPs and TIMPs fluctuations disappears upon removal of EGFR (Fig. 6). Therefore, in the presence of EGFR, S100A4's activity stabilizes the state of reduced capillary growth (corresponding to low activity of MMPs, see Fig. 5D and 6A). This suggests a positive contribution of S100A4 to the stability of a physiological phenotype in the presence of functional EGFR. Conversely, high S100A4 makes high angiogenic regimes less sensitive to MMPs and TIMPs in the network with inhibited EGFR (Fig. 5BversusFig. 5D). This shows that S100A4 can have opposite effects on the network dynamics upon inactivation of a single network component. The inactivation of EGFR-mediated feedback of S100A4 is sufficient to reverse the effect of S100A4 turning it into a stabilizing factor for low angiogenic regimes without modifying the corresponding steady-state activity profile. In addition, the described near-zero, branched sensitivity pattern shown in Fig. 6 supports potential multistable states of the system due to fluctuations of MMPs basal activity. The system can assume intermediate MMPs activity ranges bridging the two extreme regions of the MMPs activity ranges therefore facilitating (stochastic) transitions between controlled and metastatic regimes. Tumour cells containing different concentrations of proteolytically active MMPs may therefore serve as basis for the emergence of cell heterogeneity. This effect is also observable in the PCA of the steady-state activity where a variable cluster including S100A4 and EGFR is most closely associated to capillary growth at intermediate S100A4 activity (Fig. 4 of the companion article30). Similar variable clusters are observed in the absence of EGFR where, however, different activity of S100A4 does not affect their relative distances: the pattern is nearly identical over the different activity ranges of S100A4 (Fig. ESI 8 (ESI†)). PCA of sensitivity values also indicates impairment of the co-regulation between S100A4 and capillary growth in the absence of EGFR.4.2 Simulations of targeted treatment
The network model was investigated in a therapeutic-oriented manner by selectively removing or inhibiting components based on the existing biological knowledge. Quantitative information from simulated knockout, targeted inhibition and their combined effect enabled the characterization of potential targets for therapeutic intervention and mechanisms leading to drug resistance. We therefore investigated the effect of blocking MMPs and/or S100A4 together with methodical variation of key network components such as EGFR, S100A4 and NF-κB. While S100A4 knockout accentuated the effect on the cell dissociation sensitivity landscape observed earlier by reduced S100A4 activity in the intact network, capillary growth sensitivity followed instead a similar trend as with EGFR inhibition. This confirmed the role of the EGFR-mediated positive feedback loop mentioned in “EGFR-mediated feedback of S100A4 is responsible for multistable buffering patterns” in co-regulating S100A4 and EGFR. In addition, across a higher sensitivity barrier with respect to the intact network, the system devoid of S100A4 was still susceptible to assume pathological regimes due to a very high activity of EGFR. This reveals the presence of a compensation mechanism that drives the system to a similar behaviour as in the presence of S100A4. It remains to be seen if high levels of active EGFR are indeed observed in tumours, or if tumours devoid of S100A4 and treated by a combination of MMPs and EGFR inhibitors fail to form metastases. Furthermore, when inhibition of MMPs was simulated, the system revealed complete insensitivity to TIMPs on the one hand and steady-state alteration of cell dissociation and capillary growth with respect to the intact network on the other. High activity of S100A4 has in this case too the potential to drive the system beyond a sensitivity barrier determined by EGFR to pathological regimes and consequently compensate for inhibited MMPs. Combination of S100A4 knockout with inhibition of MMPs further impairs both cell dissociation and capillary growth through barriers in the sensitivity landscape that increase sensitivity to EGFR's activity and at the same time decrease the steady-state levels of cell dissociation.Interestingly, network modifications involving EGFR-mediated S100A4 translocation did not significantly alter the steady-state levels of cell dissociation and capillary growth but had a strong effect on their sensitivity profile. This effect was marked by an overall increase of the relative sensitivity barrier separating low from high activity levels. In contrast, modifications involving MMPs (inhibited alone or in combination with other targets) resulted in a decrease of cell dissociation and an increase of capillary growth steady-state with respect to the intact network.
5 Conclusions
We used a steady-state simulation method to study a network leading to metastasis in solid cancers (breast cancer in particular), where the key interactions involve S100A4, various MMPs and their tissue inhibitors, EGFR and NF-κB. Through selective knockout or inhibition of the network components, we have studied potential therapeutic interventions. Our results suggest that while MMP inhibitors are not expected to be useful in tumours that express S100A4, they can otherwise be useful as inhibitors of metastasis as long as the concentration of active EGFR in the cells is not too high. Other potential therapies have also been discussed. Overall, our approach is useful to decide on experiments that involve regulation of biological networks.Acknowledgements
We acknowledge Prof. Gunhild M. Mælandsmo together with her staff for helpful discussions and Kirill Lykov for technical help with computational infrastructures. The calculations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at Lunarc, centre for scientific and technical computing for research at Lund University and by the Swiss National Supercomputing Centre (CSCS). I.V.P. acknowledges the support from Swiss National Science Foundation and Swiss Platform for Advanced Scientific Computing. This work was supported by Holcim Foundation (Switzerland, to AB) and Crafoord Foundation (Sweden, grant application numbers 20120856 and 20130787, to RF).References
- L. Santamaria-Kisiel, A. C. Rintala-Dempsey and G. S. Shaw, Biochem. J., 2006, 396(2), 201–214 CrossRef CAS PubMed.
- I. Salama, P. S. Malone, F. Mihaimeed and J. L. Jones, Eur. J. Surg. Oncol., 2008, 34(4), 357–364 CrossRef CAS PubMed.
- L. J. Sparvero, D. Asafu-Adjei, R. Kang, D. Tang, N. Amin, J. Im, R. Rutledge, B. Lin, A. A. Amoscato, H. J. Zeh and M. T. Lotze, J. Transl. Med., 2009, 7, 17 CrossRef PubMed.
- H. Chen, C. Xu, Q. Jin and Z. Liu, Am. J. Cancer Res., 2014, 4(2), 89–115 Search PubMed.
- K. Boye and G. M. Mælandsmo, Am. J. Pathol., 2010, 176(2), 528–535 CrossRef CAS PubMed.
- T. Cabezón, J. E. Celis, I. Skibshoj, J. Klingelhöfer, M. Grigorian, P. Gromov, F. Rank, J. H. Myklebust, G. M. Mælandsmo, E. Lukanidin and N. Ambartsumian, Int. J. Cancer, 2007, 121, 1433–1444 CrossRef PubMed.
- K. Takenaga, H. Nakanishi, K. Wada, M. Suzuki, O. Matsuzaki, A. Matsuura and H. Endo, Clin. Cancer Res., 1997, 3, 2309–2316 CAS.
- S. Gongoll, G. Peters, M. Mengel, P. Piso, J. Klempnauer, H. Kreipe and R. von Wasielewski, Gastroenterology, 2002, 123, 1478–1484 CrossRef CAS PubMed.
- B. R. Davies, M. P. Davies, F. E. Gibbs, R. Barraclough and P. S. Rudland, Oncogene, 1993, 8, 999–1008 CAS.
- M. Grigorian, N. Ambartsumian, A. E. Lykkesfeldt, L. Bastholm, F. Elling, G. Georgiev and E. Lukanidin, Int. J. Cancer, 1996, 67, 831–841 CrossRef CAS.
- M. Fujiwara, T. G. Kashima, A. Kunita, I. Kii, D. Komura, A. E. Grigoriadis, A. Kudo, H. Aburatani and M. Fukayama, Tumour Biol., 2011, 611–622 CrossRef CAS PubMed.
- T. Tabata, N. Tsukamoto, A. A. Fooladi, S. Yamanaka, T. Furukawa, M. Ishida, D. Sato, Z. Gu, H. Nagase, S. Egawa, M. Sunamura and A. Horii, Biochem. Biophys. Res. Commun., 2009, 390, 475–480 CrossRef CAS PubMed.
- R. R. Bowers, Y. Manevich, D. M. Townsend and K. D. Tew, Biochemistry, 2012, 51(39), 7740–7754 CrossRef CAS PubMed.
- S. Tarabykina, T. R. Griffiths, E. Tulchinsky, J. K. Mellon, I. B. Bronstein and M. Kriajevska, Curr. Cancer Drug Targets, 2007, 7, 217–228 CrossRef CAS.
- Z. H. Li and A. R. Bresnick, Cancer Res., 2006, 66(10), 5173–5180 CrossRef CAS PubMed.
- J. Zhang, D. L. Zhang, X. L. Jiao and Q. Dong, Eur. Rev. Med. Pharmacol. Sci., 2013, 17(17), 2372–2382 CAS.
- W. Jia, X. J. Gao, Z. D. Zhang, Z. X. Yang and G. Zhang, Eur. Rev. Med. Pharmacol. Sci., 2013, 17(11), 1495–1508 CAS.
- M. Saleem, M. H. Kweon, J. J. Johnson, V. M. Adhami, I. Elcheva, N. Khan, B. Bin Hafeez, K. M. Bhat, S. Sarfaraz, S. Reagan-Shaw, V. S. Spiegelman, V. Setaluri and H. Mukhtar, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(40), 14825–14830 CrossRef CAS PubMed.
- N. Ambartsumian, J. Klingelhöfer, M. Grigorian, C. Christensen, M. Kriajevska, E. Tulchinsky, G. Georgiev, V. Berezin, E. Bock, J. Rygaard, R. Cao, Y. Cao and E. Lukanidin, Oncogene, 2001, 20(34), 4685–4695 CrossRef CAS PubMed.
- A. Semov, M. J. Moreno, A. Onichtchenko, A. Abulrob, M. Ball, I. Ekiel, G. Pietrzynski, D. Stanimirovic and V. Alakhov, J. Biol. Chem., 2005, 280, 20833–20841 CrossRef CAS PubMed.
- S. de Silva Rudland, L. Martin, C. Roshanlall, J. Winstanley, S. Leinster, A. Platt-Higgins, J. Carroll, C. West, R. Barraclough and P. Rudland, Clin. Cancer Res., 2006, 12, 1192–1200 CrossRef CAS PubMed.
- G. Berge, S. Pettersen, I. Grotterød, I. J. Bettum, K. Boye and G. M. Mælandsmo, Int. J. Cancer, 2011, 129, 780–790 CrossRef CAS PubMed.
- M. Schneider, J. L. Hansen and S. P. Sheikh, J. Mol. Med., 2008, 86(5), 507–522 CrossRef CAS PubMed.
- K. Bjrnland, J. O. Winberg, O. T. Odegaard, E. Hovig, T. Loennechen, A. O. Aasen, O. Fodstad and G. M. Mælandsmo, Cancer Res., 1999, 59(18), 4702–4708 Search PubMed.
- J. Klingelhöfer, H. D. Møller, E. U. Sumer, C. H. Berg, M. Poulsen, D. Kiryushko, V. Soroka, N. Ambartsumian, M. Grigorian and E. M. Lukanidin, FEBS J., 2009, 276(20), 5936–5948 CrossRef PubMed.
- R. Hernan, R. Fasheh, C. Calabrese, A. J. Frank, K. H. Maclean, D. Allard, R. Barraclough and R. J. Gilbertson, Cancer Res., 2003, 63(1), 140–148 CAS.
- M. Grigorian, S. Andresen, E. Tulchinsky, M. Kriajevska, C. Carlberg, C. Kruse, M. Cohn, N. Ambartsumian, A. Christensen, G. Selivanova and E. Lukanidin, J. Biol. Chem., 2001, 276(25), 22699–22708 CrossRef CAS PubMed.
- G. Berge and G. M. Mælandsmo, Amino Acids, 2011, 41(4), 863–873 CrossRef CAS PubMed.
- L. M. Orre, E. Panizza, V. O. Kaminskyy, E. Vernet, T. Gräslund, B. Zhivotovsky and J. Lehtiö, Oncogene, 2013, 32(49), 5531–5540 CrossRef CAS PubMed.
- A. Buetti-Dinh, I. V. Pivkin and R. Friedman, Mol. BioSyst. Search PubMed , under review.
- E. Aréchaga-Ocampo, N. Villegas-Sepulveda, E. Lopez-Urrutia, M. Ramos-Suzarte, C. Lopez-Camarillo, C. Perez-Plasencia, C. H. Gonzalez-de la Rosa, C. Cortes-Gonzalez and L. A. Herrera, Biomarkers in Lung Cancer: Integration with Radiogenomics Data, in Oncogenomics and Cancer Proteomics – Novel Approaches in Biomarkers Discovery and Therapeutic Targets in Cancer, ed. C. Lopez, 2013 ISBN: 978-953-51-1041-5, InTech, DOI:10.5772/53426, http://www.intechopen.com/books/oncogenomics-and-cancer-proteomics-novel-approaches-in-biomarkers-discovery-and-therapeutic-targets-in-cancer/biomarkers-in-lung-cancer-integration-with-radiogenomics-data.
- T. G. Whitsett, A. Richer, J. Friel, V. Carson and L. Inge, Pharmacogenomics Pers. Med., 2015, 63 CrossRef PubMed.
- D. Hanahan and R. A. Weinberg, Cell, 2000, 100, 57–70 CrossRef CAS.
- K. Hirota, M. Murata, T. Itoh, J. Yodoi and K. Fukuda, J. Biol. Chem., 2001, 276, 25953–25958 CrossRef CAS PubMed.
- A. A. Habib, S. Chatterjee, S. K. Park, R. R. Ratan, S. Lefebvre and T. Vartanian, J. Biol. Chem., 2001, 276, 8865–8874 CrossRef CAS PubMed.
- S. S. Sidhu, R. Nawroth, M. Retz, H. Lemjabbar-Alaoui, V. Dasari and C. Basbaum, Oncogene, 2010, 29, 4145–4156 CrossRef CAS PubMed.
- K. Boye, J. M. Nesland, B. Sandstad, M. Haugland Haugen, G. M. Mælandsmo and K. Flatmark, Br. J. Cancer, 2012, 107, 667–674 CrossRef CAS PubMed.
- S. Sarkar, R. Swiercz, C. Kantara, K. A. Hajjar and P. Singh, Gastroenterology, 2011, 140, 583–595 CrossRef CAS PubMed.
- R. Albert, H. Jeong and A. L. Barabasi, Nature, 2000, 406, 378–382 CrossRef CAS PubMed.
- H. Jeong, B. Tombor, R. Albert, Z. N. Oltvai and A. L. Barabasi, Nature, 2000, 407, 651–654 CrossRef CAS PubMed.
- V. N. Malashkevich, N. G. Dulyaninova, U. A. Ramagopal, M. A. Liriano, K. M. Varney, D. Knight, M. Brenowitz, D. J. Weber, S. C. Almo and A. R. Bresnick, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(19), 8605–8610 CrossRef CAS PubMed.
- U. Sack, W. Walther, D. Scudiero, M. Selby, J. Aumann, C. Lemos, I. Fichtner, P. M. Schlag, R. H. Shoemaker and U. Stein, Mol. Biol. Cell, 2011, 22(18), 3344–3354 CrossRef CAS PubMed.
- B. Schmidt-Hansen, D. Orns, M. Grigorian, J. Klingelhöfer, E. Tulchinsky, E. Lukanidin and N. Ambartsumian, Oncogene, 2004, 23(32), 5487–5495 CrossRef CAS PubMed.
- L. Salvatori, F. Caporuscio, A. Verdina, G. Starace, S. Crispi, M. R. Nicotra, A. Russo, R. A. Calogero, E. Morgante, P. G. Natali, M. A. Russo and E. Petrangeli, PLoS One, 2012, 7(2), e31467 CAS.
- C. M. Overall and O. Kleifeld, Nat. Rev. Cancer, 2006, 6(3), 227–239 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mb00302d |
| This journal is © The Royal Society of Chemistry 2015 |