 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Naïve Bayes classifier predicts functional microRNA target interactions in colorectal cancer†
Raheleh
Amirkhah
a,
Ali
Farazmand‡
*a,
Shailendra K.
Gupta
bc,
Hamed
Ahmadi
d,
Olaf
Wolkenhauer
be and
Ulf
Schmitz§
*b
aDepartment of Cell and Molecular Biology, School of Biology, College of Science, University of Tehran, Tehran, Iran
bDepartment of Systems Biology and Bioinformatics, Institute of Computer Science, University of Rostock, Rostock, Germany
cDepartment of Bioinformatics, CSIR-Indian Institute of Toxicology Research, Lucknow, India
dMultimedia Processing Laboratory (MPL), School of Electrical and Computer Engineering, College of Engineering, University of Tehran, Tehran, Iran
eStellenbosch Institute for Advanced Study (STIAS), Wallenberg Research Centre at Stellenbosch University, Stellenbosch, South Africa
First published on 11th June 2015
Abstract
Alterations in the expression of miRNAs have been extensively characterized in several cancers, including human colorectal cancer (CRC). Recent publications provide evidence for tissue-specific miRNA target recognition. Several computational methods have been developed to predict miRNA targets; however, all of these methods assume a general pattern underlying these interactions and therefore tolerate reduced prediction accuracy and a significant number of false predictions. The motivation underlying the presented work was to unravel the relationship between miRNAs and their target mRNAs in CRC. We developed a novel computational algorithm for miRNA–target prediction in CRC using a Naïve Bayes classifier. The algorithm, which is referred to as CRCmiRTar, was trained with data from validated miRNA target interactions in CRC and other cancer entities. Furthermore, we identified a set of position-based, sequence, structural, and thermodynamic features that identify CRC-specific miRNA target interactions. Evaluation of the algorithm showed a significant improvement of performance with respect to AUC, and sensitivity, compared to other widely used algorithms based on machine learning. Based on miRNA and gene expression profiles in CRC tissues with similar clinical and pathological features, our classifier predicted 204 functional interactions, which involve 11 miRNAs and 41 mRNAs in this cancer entity. While the approach is here validated for CRC, the implementation of disease-specific miRNA target prediction algorithms can be easily adopted for other applications too. The identification of disease-specific miRNA target interactions may also facilitate the identification of potential drug targets.
Introduction
In recent years, many publications have highlighted the functional role of microRNAs (miRNA) in CRC.1 MiRNAs are small non-coding RNA molecules of about ∼22 nucleotides in length which have critical functions across various biological processes.2 They act by binding to complementary sites in the 3′ untranslated region (UTR) of their target genes to either induce degradation of the target transcript, or to repress its translation into a protein.3 MiRNAomics studies have detected dysregulation of miRNAs in the broad spectrum of haematological malignancies and solid tumours, including CRC.4 Experimental detection of miRNA targets is a costly and time-consuming process and likewise the experimental investigation of miRNA-induced consequences for signalling pathways and cellular function.5 An efficient detection of novel miRNA target interactions benefits from reliable computational predictions. However, there is still room for improvement with respect to specificity in the established generic algorithms. The identification of tissue and disease-specific miRNA target genes would ultimately contribute to the understanding of their biological functions.Hence, the development of computational methods for miRNA target prediction is fundamental for understanding the role of miRNAs in gene regulation. To date various packages available that can predict miRNA targets in mammals. Most of these algorithms are based on similar principles for the identification of putative target sites in mRNA 3′ UTR sequences, which include: (i) sequence complementarity between miRNA and target site (with focus on the seed region), (ii) target site conservation in related species, (iii) thermodynamic stability of a miRNA–mRNA duplex, and (iv) site accessibility.6 Computational approaches for miRNA target prediction can be classified in two main categories: ab initio methods and machine learning (ML)-based approaches. While ab initio target prediction is based on empirical evidence with respect to binding patterns, ML-based approaches benefit from statistically derived patterns in sequence, structure and loci. Therefore, ML-based approaches where established at the time when a statistically significant number of miRNA–target pairs were known. These algorithms are able to reduce the high number of false positive predictions of ab initio methods.7 Though a couple of studies have applied ML methods, the rate of false positive predictions is still an issue of concern, which may be due to the tissue and disease specificity in miRNA regulation.
Since no gold standard training dataset exists, the developers of ML-based algorithms have tested their methods on different data. Most of these algorithms use data from miRTarbase8 and TarBase,9 two databases of experimentally confirmed miRNA target interactions, for training. However, details on the miRNA binding sites in their respective targets are often missing in these databases. Therefore, different miRNA target prediction algorithms generate differing results, and often researchers tend to consider only those predictions that are common among multiple algorithms in order to have an additional layer of confidence on predicted targets.10 Thereby they may however loose valid interactions that are not part of the intersection set.
Recently, Clark et al. demonstrated miRNA targetome diversity across tissue types by analysing Argonaute CLIP-Seq data.11 They analyzed 34 Argonaute HITS-CLIP datasets from several human and mouse cell types and discovered that many miRNA–target heteroduplexes are non-canonical, i.e. their seed region comprises G:U wobble pairs and bulges, while most of the current algorithms consider perfect 6mer, 7mer and 8mer seed matches only.11 Hence, the reliable prediction of a functional miRNA target in a tissue-specific manner is still a challenging task. Based on the highly tissue-specific expression signatures of miRNAs and target transcripts, tissue-specific miRNA function has to be considered to improve the analysis of miRNA regulation under specific pathological conditions. In a recent publication, Bandyopadhyay et al. reported that all predicted miRNA targets using current computational approach are not functional in all tissues or diseases.12 In fact some binding sites of previously validated targets were not accessible for miRNA binding in another tissue because they are occluded by the mRNA secondary structure or masked by RNA binding proteins.13,14
Fortunately, with a sufficient amount of data on miRNAs and their targets available, it is now possible to develop computational methods that can effectively predict disease-specific miRNA targets.
In this work, we present a reliable model for the prediction of miRNA–target interactions specific to CRC. For this purpose we trained a ML-based classifier with data from experimentally validated miRNA target sites in CRC cells. ML-based algorithms are data-driven, i.e. the dataset used for training has a high impact on the classification performance. Therefore, we applied two strict filters in the data selection step to ensure the reliability of our dataset: (i) the data should be experimentally validated for CRC; and (ii) the exact binding site should have been identified by luciferase reporter or mutagenesis assays. The data consists of sequence, structure, thermodynamic and position-based features extracted from the experimental results. These features represent a collection of features used in other generic target prediction algorithms including TargetSpy15 and MultimiTar16 with an emphasis on sequence-related features.
In addition, we applied two feature selection methods to identify a subset of most relevant features. We compared the classification performance of several ML methods (Naïve Bayes (NB), Random forest (RF), Artificial Neural Network (ANN), Support Vector Machine (SVM)) based on which we decided to establish a NB classifier to unravel the interactions between miRNAs and target mRNAs in CRC. This classifier we refer to as CRCmiRTar. Fig. 1 shows the workflow implemented here. Evaluation of the classifier showed a significant improvement of performance with respect to AUC, and sensitivity, compared to other widely used machine learning-based algorithms. Based on miRNA and gene expression profiles in patient-derived CRC tissue samples with similar clinical and pathological features our classifier predicted 204 functional interactions which involve 11 miRNAs and 41 mRNAs in this cancer entity. These results can be accessed in Table S1 (ESI†).
 | ||
| Fig. 1 Implementation of the CRCmiRTar workflow. The positive dataset (miRNA–mRNA interactions in CRC) contains literature-based experimentally validated interactions. The negative dataset consists of the tissue-specific negative data which was previously used in training the TargetMiner algorithm21 and validated miRNA–target interactions from other cancers based on the miR2Disease database. In the negative dataset, for uncharacterised binding sites, we used the Smith–Waterman algorithm to localize the binding sites. In the next step, those interactions which were common between CRC and other cancers were deliminated from the negative training data. In order to establish a classifier, first, 70 features which are used in previous studies were extracted from all positive and negative interactions. Next, two different feature selection methods were used to select the most informative features. We assessed the performance of the different classifiers based on 10-fold cross-validation and an independent test dataset. | ||
Results and discussion
Structural features determine CRC-specificity
A key step in the identification of miRNA targets is the selection of features that have strong predictive power. We applied CFS and ReliefF to identify optimal features for our machine learning classifier. The best performing subset that was identified by CFS on the training dataset contains the 14 features listed in Table 1. The presence of nine structural features among the selected features suggests that the structural layout of a putative miRNA–mRNA hybrid is a predominant determinant of whether this hybrid is functional in CRC or not. These nine structural features are: (i–iv) frequency of base pairs between miRNA and mRNA (A:U, U:A, G:C, and C:G); (v) the number of matches in the seed site; (vi) number of matches in the miRNA tail (last eight nucleotides of the miRNA); (vii) consecutive base pairings in the miRNA 3′ end (with two non-pairing positions allowed); (viii) binding asymmetry (ratio between the number of paired bases in the 3′ vs. the 5′ region of the miRNA); and (ix) number of bulges of size 6 nt or more in the target site.| Features | Description | Feature type |
|---|---|---|
| UU_seed | UU's frequency in seed matching site | Sequence |
| CG_seed | CG's frequency in seed matching site | Sequence |
| AU_match | Frequency of AU base pair in seed region | Structural |
| UA_match | Frequency of UA base pair in seed region | Structural |
| GC_target | GC's frequency in target site | Sequence |
| GC_match | Frequency of GC base pair in seed region | Structural |
| CG_match | Frequency of CG base pair in seed region | Structural |
| Seed | Number of base pairings to the miRNA 8-mer seed | Structural |
| Tail | Number of base parings to the first 8 nucleotides of the miRNA 3′ end | Structural |
| Cons_bp_mir_5p | Number of consecutive base-pairings to the miRNA 5′ end with two allowed non-pairing positions | Structural |
| Binding asymmetry | The ratio between the number of paired bases in the 3p versus the 5p region of the microRNA (considering 8 nucleotides on each side) | Structural |
| B_tagt_s6 | Bulges in target sequences of size 6 nt and more | Structural |
| Pos_3 | Position 3 | Position |
| Pos_7 | Position 7 | Position |
The features (vi–vii) indicate the importance of the 3′ part of the miRNA for the stability of the miRNA–mRNA duplex. The frequencies of the (di-) nucleotides UU and CG in the seed and frequency of G and C in the target site are some of the sequence-based features that appear in the optimal feature set identified by CFS. The two remaining features are position-based features that focus on the matching type in the positions 3, and 7 of the seed region. Compared to the 14 CFS-selected features, the top 14 ranking features from ReliefF have nine features in common with those selected by CFS (see Table S2, the common features are in red, ESI†), the others are: the GC dinucleotide frequency in the seed, matching type in positions 2, 4 and 5. Interestingly, the minimum free energy (MFE) of the duplex was not identified as important in the feature selection process although the energy was previously shown to have an impact target repression efficacy.17 The reason for this observation is that both the positive and the negative training data contain cases of functional miRNA–target interaction, however, some are specific to CRC and others are associated with different cancers. Therefore, we conclude that instead of the energy, CRC specificity in miRNA–target regulation is mainly based on structural features.
Naïve Bayes classifier performs best on independent test set
Before we decided to use a particular approach we compared six machine learning methods: NB, RF, ANN, and SVM with linear kernel function and non-linear kernel function. All methods have gone through 10-fold cross-validation using (i) the selected features from CFS, and (ii) the 16 top ranked features from ReliefF. Grid-based search tools provided by the LibSVM library were used to select the optimal values for parameters C and g for SVM from the training datasets.Fig. 2 shows the performance of the six methods after 10 fold cross validation by computing two different performance metrics: (i) area under the receiver operating characteristic curve (AUC) which is used to illustrate the specificity–sensitivity trade-off, and (ii) sensitivity. As it can be seen in Fig. 2a, regarding AUC, for both features sets Naïve Bayes shows the best result (AUC = 0.957), while in Fig. 2b, in terms of sensitivity, the Naïve Bayes classifier trained with the CFS selected features achieves the highest value (AUC = 0.93). Therefore, we decided to use the Naïve Bayes algorithm trained with the CFS selected features as classifier for the prediction of CRC-specific miRNA–target interactions. We name this novel classifier as CRCmiRTar.
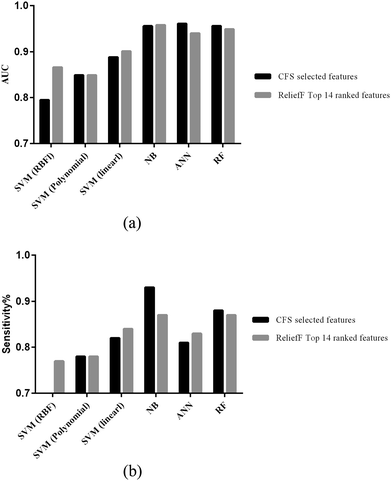 | ||
| Fig. 2 Evaluation of different classifiers on two categories of selected features using CFS and ReliefF. These plots illustrate the performances of the different classifiers in the 10-fold cross validation: (a) AUC; (b) sensitivity values. Results of the re-evaluation step with a separate test dataset can be found in Fig. S1 (ESI†). RBF: radial basis function kernel. | ||
Structural features are important in determining CRC-specificity
We analysed the contribution of each type of feature, i.e. position-based, sequence and structural features, among the selected features from CFS to the performance of CRCmiRTar. The performance of the classifier was evaluated based on the 10-fold cross validation using ROC curves (see Fig. 3). The plot illustrates the specificity–sensitivity trade-off, i.e. the true positive rate against the false positive rate. Using all CFS selected features (rectangular) resulted in an AUC of 95.7%. For structural features we observed a slight AUC reduction to 93.6% without any significant effect in sensitivity of the classifier (circle). In case of using only sequence features the AUC drops to 67.5% (plus). Finally, when considering only position-based features the AUC further decreases to 68.4% (triangle). In addition, we evaluated the performance by combining any two types of features, i.e. structural and sequence features, structural and position-based features as well as sequence and position-based features. We found that the combination of structural and position-based features results in the highest value for the area under the ROC curve (AUC = 0.954). These results indicate that structural features are important in determining CRC-specificity, however the combination of all, the structural, sequence, and position-based features is necessary to achieve an optimal performance in the classification of CRC-specific miRNA–target interactions. Table 2 shows the sensitivity and specificity of the model for each type of feature and combination of them. As can be seen the structural features ensure a high sensitivity of the model, while the sequence features contribute towards the high specificity of CRCmiRTar.| Sensitivity | Specificity | F-measure | AUC | |
|---|---|---|---|---|
| All selected Features | 0.93 | 0.861 | 0.883 | 0.956 |
| Structural features | 0.94 | 0.813 | 0.853 | 0.936 |
| Sequence features | 0.27 | 0.912 | 0.68 | 0.675 |
| Position-based features | 0.50 | 0.70 | 0.67 | 0.684 |
| Structural + sequence | 0.92 | 0.84 | 0.86 | 0.938 |
| Structural + position | 0.94 | 0.83 | 0.872 | 0.954 |
| Sequence + position | 0.58 | 0.81 | 0.756 | 0.803 |
Additionally, to test if the same features would be selected in another cancer; we tested our methodology for breast cancer (66 positive samples) and lung cancer (70 positive samples) specific miRNA target interactions. The CFS-based feature selection resulted in largely different sets of features (in number as well as in type) that seem to be relevant for these cancer types. This emphasizes the necessity to re-perform the whole analysis for each disease individually in order to obtain a customized disease-specific set of features that are able to reliably predict miRNA–target interactions functional in this disease. We included the comparison in Table S3 (ESI†).
CRCmiRTar more sensitive than other related tools
We compared the performance of CRCmiRTar, with MultiMiTar16 and RFMirTarget18 which are both recent and well performing algorithms and with miTarget which was the first miRNA target prediction method based on a ML approach. MultiMiTar is a SVM based classifier integrated with a novel feature selection technique, AMOSA-SVM. In their publication the authors were showing that MultiMiTar outperforms many other well-known target prediction methods. RFMirTarget is a recent algorithm based on a random forest approach that could outperform MultiMiTar and several other well-known classifiers.To make a comparison, we re-implemented these algorithms and trained them with the same data as was used to train our model. Results of the comparison regarding sensitivity, specificity, and Matthew's correlation coefficient (MCC), which is the quality measure of a binary classification, are shown in Table 3. In terms of MCC, CRCmiRTar (MCC = 0.726) shows a ∼14% and ∼6% increase compared to miTarget and RFMirTarget, respectively. CRCmiRTar provides the highest sensitivity among the four predictors (0.93), which is a ∼16% increase to the second best performing classifier, MultiMiTar (0.77). The specificity of our model is a little lower than that of the others (0.86). Even though the specificity is marginally better for the other tools, their sensitivity is remarkably reduced and as a result there is disequilibrium in their performance. Instead, CRCmiRTar provides the most balanced result in terms of sensitivity and specificity as compared to the others which is underlined by its high AUC value (0.957) compared to MultiMiTar (0.943), RFMirTarget (0.92) and miTarget (0.884). In addition, the ROC curves plotted in Fig. 4 confirm the effectiveness of CRCmiRTar in discriminating between functional and non-functional miRNA–mRNA interactions in CRC. Interestingly, the common features between our model and MultiMiTar as the second best performing model show again that sequence and structural features are very important in CRC. About half of the features in miTarget and RFMirTarget are related to thermodynamic and position-based features. As we have already shown before, these types of features are less suitable for a reliable prediction of CRC-specific miRNA targets. This may explain why miTarget and RFMirTarget perform worse in terms of sensitivity and the AUC.
| Sensitivity | Specificity | MCC | AUC | |
|---|---|---|---|---|
| CRCmiRTar | 0.93 | 0.86 | 0.726 | 0.957 |
| MultiMiTar | 0.77 | 0.965 | 0.77 | 0.943 |
| miTarget | 0.63 | 0.922 | 0.581 | 0.884 |
| RFMirTarget | 0.69 | 0.942 | 0.666 | 0.92 |
CRCmiRTar outperformes established algorithms on independent test data
We investigated the power of our model in predicting experimentally validated CRC-specific miRNA–mRNA interactions in comparison with previous algorithms. To this end, we collected 47 wet lab validated miRNA–target pairs for CRC from recently published papers and OncomiRDB which are not included in our training dataset and used the data to evaluate our method along with six other commonly employed miRNA target-prediction methods, including MirTarget2,19 TargetMiner,20 PicTar,21 TargetSpy,15 SVMicro,22 and TargetScan.23 As shown in Table 4, TargetScan and SVMicro correctly identified 32 and 27 miRNA–target pairs respectively and thus performed better than any of the other previous methods. However, CRCmiRTar could identify 41 miRNA–target pairs, which comprises more than 87% of all cases. A reason for this improvement can be the combination of features and CRC specific training dataset that we used in CRCmiRTar. An important advantage in our model training procedure compared to the strategies in other algorithms is that we consider only reliably validated miRNA–target interactions from luciferase reporter assay and site-directed mutagenesis experiments.| miRNA | Target | PMID | TargetMiner | MirTarget2 | TargetSpy | SVMicro | TargetScan | PicTar | CRCmiRTar |
|---|---|---|---|---|---|---|---|---|---|
| miR-17-5p | PTEN | 24912422 | ✓ | — | — | ✓ | ✓ | ✓ | ✓ |
| miR-139-5p | NOTCH1 | 25149074 | — | ✓ | ✓ | — | ✓ | ✓ | ✓ |
| miR-455-5p | RAF1 | 25355599 | — | — | — | — | ✓ | — | ✓ |
| miR-18a-5p | CDC42 | 25379703 | ✓ | — | — | — | — | ✓ | ✓ |
| miR-29c-3p | GNA13 | 25193986 | — | — | — | — | ✓ | ✓ | ✓ |
| miR-133b | TBPL1 | 24870791 | — | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| miR-182-5p | SATB2 | 24884732 | ✓ | ✓ | — | ✓ | — | ✓ | ✓ |
| miR-185 | STIM1 | 25531324 | — | — | — | — | — | — | ✓ |
| miR-301a | SOCS6 | 25591765 | ✓ | — | ✓ | ✓ | ✓ | ✓ | ✓ |
| miR-150 | MYB | 25230975 | — | ✓ | ✓ | ✓ | ✓ | — | ✓ |
| miR-143-3p | TLR2 | 23866094 | — | ✓ | — | — | ✓ | — | ✓ |
| miR-150-5p | MUC4 | 25124610 | — | — | — | — | ✓ | — | ✓ |
| miR-133a | FSCN1 | 25621061 | — | — | ✓ | ✓ | ✓ | — | ✓ |
| miR-16-5p | BIRC5 | 23380758 | — | — | — | ✓ | — | — | ✓ |
| miR-21-5p | TGFBR2 | 22072622 | ✓ | — | — | ✓ | ✓ | — | — |
| miR-145-5p | PAK4 | 22766504 | — | — | — | — | — | ✓ | ✓ |
| miR-137 | PXN | 23275153 | — | — | — | — | ✓ | ✓ | ✓ |
| miR-126-3p | IRS1 | 24312276 | — | — | — | — | ✓ | ✓ | ✓ |
| miR-135b-5p | MTSS1 | 24343340 | — | — | — | ✓ | ✓ | ✓ | — |
| miR-154-5p | TLR2 | 24242044 | — | — | — | ✓ | ✓ | — | ✓ |
| miR-137 | FMNL2 | 20473940 | ✓ | ✓ | — | ✓ | ✓ | ✓ | ✓ |
| miR-137 | CDC42 | 20473940 | — | — | — | ✓ | ✓ | ✓ | ✓ |
| miR-139-5p | RAP1B | 22642900 | ✓ | — | — | — | ✓ | ✓ | ✓ |
| miR-146a-5p | MMP16 | 22348245 | ✓ | ✓ | — | ✓ | ✓ | — | ✓ |
| miR-148b-3p | CCKBR | 22020560 | ✓ | — | — | ✓ | ✓ | ✓ | ✓ |
| miR-149-5p | SP1 | 22821729 | ✓ | — | — | — | ✓ | — | ✓ |
| miR-185-5p | RHOA | 21186079 | ✓ | ✓ | — | ✓ | — | ✓ | ✓ |
| miR-185-5p | CDC42 | 21186079 | ✓ | ✓ | — | ✓ | ✓ | ✓ | ✓ |
| miR-186-5p | CSNK2A1 | 23137536 | — | — | — | ✓ | — | ✓ | ✓ |
| miR-20a-5p | BNIP2 | 21242194 | ✓ | ✓ | — | ✓ | ✓ | ✓ | ✓ |
| miR-21-5p | RHOB | 21872591 | — | — | — | ✓ | ✓ | ✓ | ✓ |
| miR-216b-5p | CSNK2A1 | 23137536 | — | — | — | ✓ | ✓ | — | ✓ |
| miR-30e-3p | HELZ | 21963845 | ✓ | ✓ | — | ✓ | ✓ | — | ✓ |
| miR-30e-3p | PIK3C2A | 21963845 | ✓ | — | — | — | — | — | ✓ |
| miR-31-5p | RASA1 | 23322774 | ✓ | ✓ | — | — | ✓ | ✓ | ✓ |
| miR-320a | NRP1 | 22134529 | — | ✓ | — | ✓ | ✓ | ✓ | ✓ |
| miR-320a | NRP1 | 22134529 | — | ✓ | — | ✓ | — | ✓ | ✓ |
| miR-337-3p | CSNK2A1 | 23137536 | — | — | — | ✓ | ✓ | — | — |
| miR-342-3p | DNMT1 | 21565830 | — | — | — | — | — | — | ✓ |
| miR-345-5p | BAG3 | 21665895 | — | — | — | — | — | — | ✓ |
| miR-491-5p | BCL2L1 | 20039318 | — | ✓ | — | ✓ | — | — | ✓ |
| miR-502-5p | RAB1B | 22580605 | — | ✓ | — | ✓ | ✓ | — | ✓ |
| miR-650 | NDRG2 | 21352815 | — | — | — | — | ✓ | — | ✓ |
| miR-7-5p | YY1 | 23208495 | — | — | — | ✓ | — | — | — |
| miR-760 | CSNK2A1 | 23137536 | — | — | — | — | ✓ | — | ✓ |
| miR-93-5p | CCNB1 | 22581829 | — | — | — | — | — | — | — |
| miR-93-5p | ERBB2 | 22581829 | — | — | — | — | — | — | — |
CRCmiRTar predicts 220 novel CRC-specific miRNA–target interactions
Using miRNA and mRNA expression profiles from eight CRC tissues and their corresponding adjacent normal tissues we used CRCmiRTar to classify miRNA–mRNA pairs with inversely correlating expression profiles. In total, CRCmiRTar classified 223 miRNA–mRNA pairs as functional interactions which are comprised of 12 miRNAs and 43 mRNAs. The maximum number of predicted targets was 30 for miR-7, and the minimum number was four for miR-224-5p. These predictions can be found in Table S1 (ESI†). We compared our prediction results with the miR2Disease24 and OncomiRDB25 databases and found three of them were already experimentally validated (with other methods than luciferase assays or site directed mutagenesis). All others are novel yet uncharacterised miRNA–target interactions. We also searched for the presence of our predicted interactions in AGO-CLIP data using the starBase database. In the collective AGO-CLIP data we found read counts for ∼26.5% of the predicted target sites. However, it has to be noted that none of these experiments has been performed in CRC tissue or corresponding cell lines.MiRNA targets are associated with cancer pathways
To obtain further insight into the biological functions of dysregulated miRNAs and their predicted targets in CRC, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.726) for the identification of overrepresentations in gene ontology (GO) terms and pathways associated with the miRNA targets. GO and pathway enrichment analysis based on the 43 differentially expressed mRNAs revealed Wnt signaling as a pathway that is significantly overrepresented in this set of genes (p < 0.005) (Table S4, ESI†). In this pathway, four genes CAMK2D, CHP2, SFRP1, and SFRP2 are downregulated in CRC.36 Interestingly, SFRP1 and SFRP2 are secreted proteins that act to inhibit Wnt activation via the Frizzled receptor. Our predictions indicate that nine and three miRNAs regulate the expression of SFRP1 and SFRP2, respectively and are thus responsible for their downregulation in CRC.Expression studies have revealed the downregulation of CAMK2D in human tumor cells. Cheng et al. deciphered that growth, migration, and proliferation of human endothelial cells were regulated by WNT5A in a CAMK2D-dependent way.27 Based on our predictions eight miRNAs are involved in the regulation of CAMK2D. Furthermore, GO enrichment analysis detected two angiogenesis related terms (vasculature development and blood vessel development) to be overrepresented in dysregulated miRNA target genes (RECK, ZFPM2, STAB2, and ARHGAP24). One of these genes, RECK, is known as a metastasis/angiogenesis suppressor gene. Our algorithm found that this gene can be regulated by four miRNAs. One of these interactions, the regulation of RECK by miR-21-5p, has been experimentally validated in CRC.28
Methods
Training data
For the purpose of building a positive dataset to train our classifier, we reviewed miRNA target identification studies related to CRC with an emphasis on experimental data from Luciferase reporter assays which is one of the most reliable methods for target identification.29 More specifically, just those miRNA–mRNA interactions for which the exact binding sites were characterized by site-directed mutagenesis were considered in the positive dataset. In this step 100 positive interactions could be retrieved, which are described in Table S5 (ESI†). Since our approach requires information on the exact target sites and this information is not always available for the interactions described in miR2Disease and the OncomiRDB we mainly relied on an in-depth literature search. In this way we also made sure that the most recent miRNA–mRNA interaction data from CRC is included in our dataset.We also searched for the presence of our selected miRNA–target interactions in AGO-CLIP data using the starBase database.30 In the collective AGO-CLIP data we found read counts for ∼70% of the target sites in our training set (in ∼30% of the cases even more than 1.000 reads). However, it has to be noted that none of these experiments has been performed in CRC tissue or corresponding cell lines.
Two kinds of negative data were collected, one set is composed of validated miRNA–mRNA interactions reported in other cancers (non CRC interactions; n = 136) and another set integrates tissue-specific negative examples that were also used as training data by TargetMiner.20 For the former set we exploited validated miRNA–mRNA interactions from other cancers such as breast and lung cancer as reported in the miR2Disease database.24 For the uncharacterised binding sites, the miRNA sequences were extracted from the miRBase database,31 and the target 3′ UTR sequences were downloaded from the Ensembl database (http://www.ensembl.org). To search for all possible alignments in each miRNA–mRNA pair, we used a Smith–Waterman local alignment algorithm and considered only those alignments with the highest score for further analysis. In the algorithm, a scoring scheme in which each G:C pair and A:U earn a score of 5 and 7 respectively, each G:U pair, a score of 1 and mismatches a score of -3, was employed. Each gap opening amounts to -8 and a gap extension is penalized with a score of -2. From the negative training data we removed those interactions which are common between CRC and other cancers in order to obtain an unambiguous dataset. We finally gathered 340 samples for the negative training dataset (see Table S6, ESI†). The dataset was split into (i) 85% for training and cross-validation, and (ii) 15% as a test set for independent evaluation.
Feature extraction from miRNA–mRNA interactions
We started our analysis with a set of 70 features which were subsequently subjected to further selection steps. In general, these features can be classified into four categories: (i) sequence features, (ii) position-based features, (iii) structural as well as (iv) thermodynamic features. In order to estimate the thermodynamic stability of a miRNA:mRNA hybrid we computed their minimum free energy (MFE) structure using RNAcofold which is part of the Vienna package.32 Structural features account for the number of matches, mismatches, G:U wobble pairs, bulges, and the stem in a miRNA:mRNA hybrid. Regarding position-based features, we assigned nominal values of 1 to 4 for each G:C match, A:U match, G:U wobble pair and mismatch in each position of seed region. Sequence features refer to the base composition of the miRNA as well as target site. Additionally we considered as a feature the miRNA–mRNAs paired expression profiles. To this end, we extracted tumor-specific miRNA and mRNA expression profiles from the NCI60 panel via the CellMiner™ database (http://discover.nci.nih.gov/cellminer/). All features are listed in Table S2 (ESI†).In order to find the features that have a dominant role in discriminating positive and negative samples, two feature selection methods were considered: (i) correlation-based feature selection (CFS)33 and (ii) ReliefF.34 While CFS is evaluating subsets of features for the correlation of individual features with the class attribute and the redundancy among the features in one set, ReliefF evaluates the goodness of a feature by repeatedly choosing a random instance and considering the value of the same feature in the nearest instance of the same and different class. The key difference between CFS and ReliefF is that CFS selects an approximately optimal subset of features, whereas ReliefF only provides a ranked list of features. The list of ranked features in CRC-specific miRNA–target interactions can be found in Table S2 (ESI†). We used the Weka 3 data mining software35 for implementation of CFS and ReliefF.
Patient derived miRNA–mRNA expression data for CRC
We retrieved a list of differentially expressed miRNAs and mRNAs from a transcriptomics and miRNAomics study in patient-derived CRC tissue samples with similar clinical and pathological features.36 Microarray expression profiles from eight CRC tissues and their corresponding adjacent normal tissues revealed 14 upregulated miRNAs and 43 downregulated mRNAs in CRC. In order to predict potential miRNA binding sites, we extracted the 3′ UTR of the upregulated mRNAs from the Ensembl database and miRNA sequences from the miRBase database31 and aligned the sequences using again the Smith–Waterman algorithm. We kept putative target sites with an alignment score S ≥ 60 for classification with CRCmiRTar.Conclusion
Many reports describe the association of miRNAs with diseases. Today, with the use of computational methods one can perform miRNA target analyses in a high-throughput manner. However, these methods often result in a large number of false positive predictions, which may not represent functional miRNA–mRNA interactions, especially in a specific disease. In fact, due to the multi-faceted nature of miRNA targeting the existing prediction algorithms cannot make perfectly reliable predictions for every pathological condition.12 Thus, it makes sense to develop a disease-specific algorithm to minimize false predictions.Although a number of studies have shown that miRNA function is tissue specific (see for example12,37) so far no study has offered an algorithm to predict miRNA targets for a specific disease.
In this study, we proposed a novel miRNA target-prediction approach specific for CRC which is based on a NB classifier and uses cancer-specific training data. In the proposed model, the use of high-quality training data in which exact binding sites are experimentally verified ensures the executing efficiency of this model, because data driven algorithms can uncover the important and real targeting characteristics from this data. Most of the existing target prediction algorithms try to provide high sensitivity with respect to the identification of true positive interactions, however, these algorithms are not designed to make out disease-specific interactions and therefore result in a high false-positive prediction rate and a low overall specificity. They are thus unreliable for the purpose of identifying disease-specific miRNA–target interactions. ML-based algorithms are data-driven, i.e. the dataset has straight impact on their performance. A careful selection of relevant features for the purpose of training is a very important determinant the performance of a machine learning algorithm. It has been shown previously that by including or discarding certain groups of features the performance of an algorithm can change drastically. For example, in Kim et al. (2005) according to the authors the sensitivity of the miTarget algorithm decreased when position-specific features where excluded. Therefore, we applied two filters in the step of preparing the training dataset: (i) we chose CRC-specific miRNA–target interactions for the positive training set, and (ii) these interaction had to be validated with luciferase assays and site-directed mutagenesis experiments. Thereby we ensured an increased specificity of our classifier. However, regarding the negative dataset we were lacking a gold standard set of negative samples. For reasons of comparability we chose a negative dataset, presented in Mitra and Bandyopadhyay (2009) that was already used in other studies.16,18 We are aware that although these data are tissue specific they may be functional in CRC as well. Therefore, as another part of our negative dataset, we used the functional data for the other cancers which are not reported to be functional in CRC.
According to the results in ref. 11, most tissue specific miRNA–mRNA interactions carry a non-canonical seed region. Therefore, in order to be able of predicting tissue specific and 3′-compensatory target sites, our model does not filter out miRNA–target site pairs with non-perfect seed matches. Additionally, some studies showed the advantage of integrating gene expression data with miRNA–target predictions. For example, Wang et al. developed a network propagation based method to infer the perturbed miRNAs and their key target genes by integrating gene expressions and global gene regulatory network information.38 Therefore, we also used miRNA–mRNAs paired expression profiles to improve the accuracy of sequence-based miRNA–target predictions. However, in the feature selection step the expression profiles were not select as part of the best performing subset of features (both using CFS and ReliefF methods).
The aim of this study was to investigate whether using CRC specific training data can help to outperform previous non tissue-specific algorithms and if so, which features are most relevant for CRC.
For the first part, our results demonstrate that compared with previous methods, CRCmiRTar could predict experimentally validated miRNA target genes with higher accuracy. Regarding the features, our results show that the sequence/base composition features have the highest contribution to the specificity of the model. Previous studies have shown that the binding sites of miRNAs have specific nucleotide and dinucleotide compositions which are significantly different between targets that are downregulated upon miRNA transfection and those that are stably expressed.19,39
Another issue regarding tissue-specific miRNA target predictions is the impact of alternative 3′ UTR isoforms, because of alternative cleavage and polyadenylation (APA). APA can lead to the potential loss of miRNA binding sites by shortening the 3′ UTR sequence of target genes.40 However, for our study no suitable data was available for deriving CRC-specific 3′ UTR isoforms. Therefore, we always considered the longest 3′ UTR annotated for each gene.
Although, the present study and some other studies demonstrate that tissue-specific miRNAs are often implicated in diseases related to a specific tissue, it remains largely unknown whether there are tissue-specific features for miRNA function. We have developed this model to serve as a useful method to obtain higher-confidence predictions for targets of miRNAs involved in CRC.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.Acknowledgements
This work was supported by the Ministry of Science, Research and Technology (MSRT) of Iran; the Research Council of the University of Tehran; and the German Federal Ministry of Education and Research (BMBF) as part of the eBio:SysMet project Grant number: 0316171.References
- R. Amirkhah, U. Schmitz, M. Linnebacher, O. Wolkenhauer and A. Farazmand, Genes, Chromosomes Cancer, 2015, 54, 129–141 CrossRef CAS PubMed.
- L. He and G. J. Hannon, Nat. Rev. Genet., 2004, 5, 522–531 CrossRef CAS PubMed.
- D. P. Bartel, Cell, 2004, 116, 281–297 CrossRef CAS.
- P. Faltejskova, M. Svoboda, K. Srutova, J. Mlcochova, A. Besse, J. Nekvindova, L. Radova, P. Fabian, K. Slaba, I. Kiss, R. Vyzula and O. Slaby, J. Cell. Mol. Med., 2012, 16, 2655–2666 CrossRef CAS PubMed.
- Y. Gusev, T. D. Schmittgen, M. Lerner, R. Postier and D. Brackett, BMC Bioinf., 2007, 8(suppl 7), S16 CrossRef PubMed.
- T. Saito and P. Saetrom, New Biotechnol., 2010, 27, 243–249 CrossRef CAS PubMed.
- P. H. Reyes-Herrera and E. Ficarra, Genomics, Proteomics Bioinf., 2012, 10, 254–263 CrossRef PubMed.
- S. D. Hsu, F. M. Lin, W. Y. Wu, C. Liang, W. C. Huang, W. L. Chan, W. T. Tsai, G. Z. Chen, C. J. Lee, C. M. Chiu, C. H. Chien, M. C. Wu, C. Y. Huang, A. P. Tsou and H. D. Huang, Nucleic Acids Res., 2011, 39, D163–D169 CrossRef CAS PubMed.
- T. Vergoulis, I. S. Vlachos, P. Alexiou, G. Georgakilas, M. Maragkakis, M. Reczko, S. Gerangelos, N. Koziris, T. Dalamagas and A. G. Hatzigeorgiou, Nucleic Acids Res., 2012, 40, D222–D229 CrossRef CAS PubMed.
- H. Zheng, R. Fu, J. T. Wang, Q. Liu, H. Chen and S. W. Jiang, Int. J. Mol. Sci., 2013, 14, 8179–8187 CrossRef PubMed.
- P. M. Clark, P. Loher, K. Quann, J. Brody, E. R. Londin and I. Rigoutsos, Sci. Rep., 2014, 4, 5947 CAS.
- S. Bandyopadhyay, D. Ghosh, R. Mitra and Z. Zhao, Sci. Rep., 2015, 5, 8004 CrossRef CAS PubMed.
- M. Kertesz, N. Iovino, U. Unnerstall, U. Gaul and E. Segal, Nat. Genet., 2007, 39, 1278–1284 CrossRef CAS PubMed.
- R. Denzler, V. Agarwal, J. Stefano, D. P. Bartel and M. Stoffel, Mol. Cell, 2014, 54, 766–776 CrossRef CAS PubMed.
- M. Sturm, M. Hackenberg, D. Langenberger and D. Frishman, BMC Bioinf., 2010, 11, 292 CrossRef PubMed.
- R. Mitra and S. Bandyopadhyay, PLoS One, 2011, 6, e24583 CAS.
- M. Rehmsmeier, P. Steffen, M. Hochsmann and R. Giegerich, RNA, 2004, 10, 1507–1517 CrossRef CAS PubMed.
- M. R. Mendoza, G. C. da Fonseca, G. Loss-Morais, R. Alves, R. Margis and A. L. Bazzan, PLoS One, 2013, 8, e70153 CAS.
- X. Wang and I. M. El Naqa, Bioinformatics, 2008, 24, 325–332 CrossRef CAS PubMed.
- S. Bandyopadhyay and R. Mitra, Bioinformatics, 2009, 25, 2625–2631 CrossRef CAS PubMed.
- A. Krek, D. Grun, M. N. Poy, R. Wolf, L. Rosenberg, E. J. Epstein, P. MacMenamin, I. da Piedade, K. C. Gunsalus, M. Stoffel and N. Rajewsky, Nat. Genet., 2005, 37, 495–500 CrossRef CAS PubMed.
- H. Liu, D. Yue, Y. Chen, S. J. Gao and Y. Huang, BMC Bioinf., 2010, 11, 476 CrossRef PubMed.
- B. P. Lewis, C. B. Burge and D. P. Bartel, Cell, 2005, 120, 15–20 CrossRef CAS PubMed.
- Q. Jiang, Y. Wang, Y. Hao, L. Juan, M. Teng, X. Zhang, M. Li, G. Wang and Y. Liu, Nucleic Acids Res., 2009, 37, D98–104 CrossRef CAS PubMed.
- D. Wang, J. Gu, T. Wang and Z. Ding, Bioinformatics, 2014, 30, 2237–2238 CrossRef CAS PubMed.
- W. Huang da, B. T. Sherman and R. A. Lempicki, Nat. Protoc., 2009, 4, 44–57 CrossRef PubMed.
- C. W. Cheng, J. C. Yeh, T. P. Fan, S. K. Smith and D. S. Charnock-Jones, Biochem. Biophys. Res. Commun., 2008, 365, 285–290 CrossRef CAS PubMed.
- M. D. Bullock, K. M. Pickard, B. S. Nielsen, A. E. Sayan, V. Jenei, M. Mellone, R. Mitter, J. N. Primrose, G. J. Thomas, G. K. Packham and A. H. Mirnezami, Cell Death Dis., 2013, 4, e684 CrossRef CAS PubMed.
- Y. Jin, Z. Chen, X. Liu and X. Zhou, Methods Mol. Biol., 2013, 936, 117–127 CAS.
- J. H. Li, S. Liu, H. Zhou, L. H. Qu and J. H. Yang, Nucleic Acids Res., 2014, 42, D92–97 CrossRef CAS PubMed.
- A. Kozomara and S. Griffiths-Jones, Nucleic Acids Res., 2014, 42, D68–D73 CrossRef CAS PubMed.
- I. L. Hofacker, Nucleic Acids Res., 2003, 31, 3429–3431 CrossRef CAS PubMed.
- M. Hall, Correlation-based feature selection for machine learning, PhD thesis, Department of Computer Science, Waikato University, New Zealand, 1999 Search PubMed.
- I. Kononenko, Estimating attributes: analysis and extensions of RELIEF, 1994 Search PubMed.
- E. Frank, M. Hall, L. Trigg, G. Holmes and I. H. Witten, Bioinformatics, 2004, 20, 2479–2481 CrossRef CAS PubMed.
- J. Fu, W. Tang, P. Du, G. Wang, W. Chen, J. Li, Y. Zhu, J. Gao and L. Cui, BMC Syst. Biol., 2012, 6, 68 CrossRef CAS PubMed.
- P. Li, X. Hua, Z. Zhang, J. Li and J. Wang, BMC Syst. Biol., 2013, 7, 112 CrossRef PubMed.
- T. Wang, J. Gu and Y. Li, BMC Bioinf., 2014, 15, 255 CrossRef PubMed.
- J. Xiao, Y. Li, K. Wang, Z. Wen, M. Li, L. Zhang and X. Guang, BMC Bioinf., 2009, 10, 427 CrossRef PubMed.
- J. W. Nam, O. S. Rissland, D. Koppstein, C. Abreu-Goodger, C. H. Jan, V. Agarwal, M. A. Yildirim, A. Rodriguez and D. P. Bartel, Mol. Cell, 2014, 53, 1031–1043 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mb00245a |
| ‡ Department of Biology, Faculty of Science, University of Tehran, Tehran, Iran. E-mail: E-mail: afarazmand@khayam.ut.ac.ir; Fax: +98 21 6405141; Tel: +98 21 6112622. |
| § Department of Systems Biology & Bioinformatics, University of Rostock, Ulmenstr. 69, 18051 Rostock, Germany. E-mail: E-mail: ulf.schmitz@uni-rostock.de; Fax: +49 381 498 7570/72; Tel: +49 381 498 7570/72. |
| This journal is © The Royal Society of Chemistry 2015 |


