 Open Access Article
Open Access ArticleBioprinting of 3D hydrogels
M. M.
Stanton
a,
J.
Samitier
bcd and
S.
Sánchez
*aef
aMax Planck Institute for Intelligent Systems, Heisenbergstr. 3, 70569 Stuttgart, Germany. E-mail: sanchez@is.mpg.de
bNanobioengineering Laboratory, Institute for Bioengineering of Catalonia (IBEC), Baldiri I Reixac 10-12, 08028 Barcelona, Spain
cThe Biomedical Research Networking Center in Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN), Maria de Luna, 11, 50018, Zaragoza, Spain
dDepartment of Electronics, University of Barcelona (UB), Martí i Franquès, 1, Barcelona 08028, Spain
eSmart nano-bio-devices Laboratory, Institute for Bioengineering of Catalonia (IBEC), Baldiri Reixac, 10-12, Barcelona 08028, Spain. E-mail: ssanchez@ibecbarcelona.eu
fInstitució Catalana de Recerca i Estudis Avançats (ICREA), Psg. Lluís Companys, 23, 08010 Barcelona, Spain
First published on 11th June 2015
Abstract
Three-dimensional (3D) bioprinting has recently emerged as an extension of 3D material printing, by using biocompatible or cellular components to build structures in an additive, layer-by-layer methodology for encapsulation and culture of cells. These 3D systems allow for cell culture in a suspension for formation of highly organized tissue or controlled spatial orientation of cell environments. The in vitro 3D cellular environments simulate the complexity of an in vivo environment and natural extracellular matrices (ECM). This paper will focus on bioprinting utilizing hydrogels as 3D scaffolds. Hydrogels are advantageous for cell culture as they are highly permeable to cell culture media, nutrients, and waste products generated during metabolic cell processes. They have the ability to be fabricated in customized shapes with various material properties with dimensions at the micron scale. 3D hydrogels are a reliable method for biocompatible 3D printing and have applications in tissue engineering, drug screening, and organ on a chip models.
Bioinks
Cell biology is influenced by chemical and physical cues of the surrounding environment.1,2 Cells cultured in 2D and 3D systems demonstrate significantly different behavior including migration, adhesion, gene expression and mitosis.3,4 3D culture constructs are important for mimicking native cell tissue in vitro5 and recently bioinks have emerged as a candidate for reliable and fast 3D bioprinted cell culture systems. Bioinks are composed of cells suspended in a liquid, pre-gel solution that is then “printed” onto a surface or into a 3D scaffold using mechanical extrusion. During the printing process, the bioink solution is gelled by polymer crosslinkers, photo activation, or thermal activation while leaving the cells intact and viable. The final bioink construct is a hydrogel that physically constrains the suspended cells. Bioink hydrogels maintain cell viability, but can be tailored for specific material properties or scaffolding dimensions.For a rapidly forming, 3D bioprinting technique, Li et al.6 combined two DNA based hydrogels for development of biodegradable bioinks (Fig. 1a). Bioink A was a polypeptide–DNA conjugate where multiple single stranded DNAs (ssDNA) were attached to a poly(L-glutamic acid240-co-γ-propargyl-L-glutamate20) (p(LGA240-co-PLG20)) backbone. Bioink B consisted of a double stranded DNA (dsDNA) with “sticky ends” consisting of ssDNA sequences complementary to the ssDNA polypeptide sequences of bioink A. A fluid 5 wt% mixture of the two bioinks with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of sticky ends in Tris–borate–EDTA buffer changed to an optically transparent crosslinked hydrogel in seconds. With a multivalve 3D bioprinter, hydrogel fabrication was performed at mm scale dimensions down to gels with 500 μm diameters and a thickness of 80 μm using 60 nL droplets. To examine the DNA-hydrogel as a 3D cell scaffold, anterior pituitary cells (AtT-20) were added to bioink A before gel polymerization. Cells remained in a homogeneous suspension within the polymerized gel and were shown to maintain normal viability and normal organelle activity indicated by fluorescent live/dead assays and high resolution organelle tracking. Long term monitoring of the cells within the hydrogel showed the cells were still viable after 48 hours of culture. To demonstrate the hydrogel as a biocompatible system, biodegradability of the hydrogel was demonstrated with a protease and nucleases. Endoproteinase Glu-C degraded the polypeptide backbone after 12 hours and the nucleases EcoRI or BamHI could cleave the DNA linkers after 24 hours. The DNA bioink system offers an advantage over synthetic polymer hydrogel systems due to its biodegradability allowing the scaffold to have medical applications as temporary scaffolding. The hydrogel is biocompatible and could be used for rapid formation of 3D constructs for tissue engineering.
1 molar ratio of sticky ends in Tris–borate–EDTA buffer changed to an optically transparent crosslinked hydrogel in seconds. With a multivalve 3D bioprinter, hydrogel fabrication was performed at mm scale dimensions down to gels with 500 μm diameters and a thickness of 80 μm using 60 nL droplets. To examine the DNA-hydrogel as a 3D cell scaffold, anterior pituitary cells (AtT-20) were added to bioink A before gel polymerization. Cells remained in a homogeneous suspension within the polymerized gel and were shown to maintain normal viability and normal organelle activity indicated by fluorescent live/dead assays and high resolution organelle tracking. Long term monitoring of the cells within the hydrogel showed the cells were still viable after 48 hours of culture. To demonstrate the hydrogel as a biocompatible system, biodegradability of the hydrogel was demonstrated with a protease and nucleases. Endoproteinase Glu-C degraded the polypeptide backbone after 12 hours and the nucleases EcoRI or BamHI could cleave the DNA linkers after 24 hours. The DNA bioink system offers an advantage over synthetic polymer hydrogel systems due to its biodegradability allowing the scaffold to have medical applications as temporary scaffolding. The hydrogel is biocompatible and could be used for rapid formation of 3D constructs for tissue engineering.
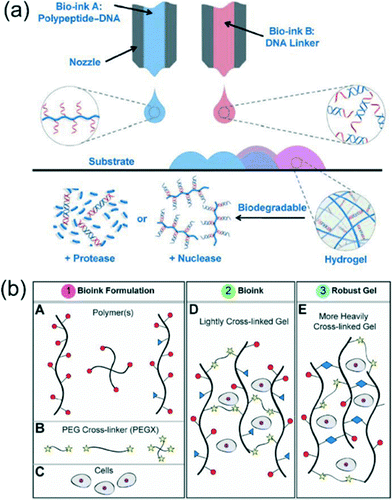 | ||
| Fig. 1 Examples of polymers and crosslinkers for bioinks. (a) 3D bioprinting of the polypeptide–DNA hydrogel to fabricate arbitrarily designed 3D structures. Bioink A (blue): polypeptide–DNA, bioink B (red): DNA linker. The DNA sequences of bioink A and bioink B are complementary, and hybridization will cause crosslinking, leading to hydrogel formation (pink). (b) (A) Polymer or polymer bioink mixtures can be linear, branched, or multifunctional. (B) PEGX can be linear or multiarm and can be various chain lengths. (C–E) Cells can be optionally incorporated by mixing with polymers and the PEGX to form the bioink. Reprinted from ref. 6 and 7 with permission from John Wiley and Sons. | ||
For a larger variety of gel-phase bioinks with tunable material properties, Rutz et al.7 devised a hydrogel fabrication method using synthetic and natural materials. A total of 35 bioink formulations were investigated in regard to their biocompatibility, bioprinting applicability, and rheological behavior. The method linked polymer solutions with a polyethylene glycol (PEG) crosslinker ending in two reactive groups (PEGX) (Fig. 1b). Polymers for the bioinks had the ability to be linear or branched and included natural proteins (gelatin and fibrinogen), modified proteins (gelatin methacrylate), synthetic polymers (PEG amine), and synthetic–natural mixtures. Bioinks were required to be extruded through a 200 μm tip for bioprinting user defined micro 3D scaffolds. By varying PEGX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer mass-to-mass ratios “soft” or “robust” gels could be identified. Robust gels were not able to be extruded through the bioink dispenser, but soft gels were capable of extrusion with continuous polymer strands ideal for building hydrogel constructs. Rheological investigations of PEGX–gelatin bioinks revealed an increase in critical stress and a decrease in critical strain when the gelatin concentration increased at a fixed PEG ratio or when the PEG concentration increased with a fixed gelatin concentration. To further modify the bioink, a secondary, postprinting crosslinking step was used to adjust the modulus and degradation properties of the gel. Printed PEGX–gelatin was further crosslinked by exposure to N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDC) and N-hydroxysuccinimide (NHS) increasing the modulus from Pa to kPa. Normal degradation of the gel occurred after 2 days, but after secondary crosslinking degradation was extended to 4 weeks. Different bioinks were successfully coprinted and were capable of directing cell adhesion. Patterning of PEGX–PEG and PEGX–gelatin regulated human dermal fibroblasts to gelatin locations. Co-cultures of human umbilical vein endothelial cells (HUVECs) and human mesenchymal stem cells (hMSCs) were spatially organized in 3D by printing PEGX–gelatin seeded with HUVECs in a micro grid and filling the open spaces with hMSCs. After two weeks the gel had degraded but cells had formed a robust tissue with the grid pattern still observable. The customizable bioinks offer synthetic and natural bioinks for a variety of tissue engineering applications and further expands the number of bioinks available for 3D printing. The tunable material properties and degradation times show promise for future cell-specific bioinks.
polymer mass-to-mass ratios “soft” or “robust” gels could be identified. Robust gels were not able to be extruded through the bioink dispenser, but soft gels were capable of extrusion with continuous polymer strands ideal for building hydrogel constructs. Rheological investigations of PEGX–gelatin bioinks revealed an increase in critical stress and a decrease in critical strain when the gelatin concentration increased at a fixed PEG ratio or when the PEG concentration increased with a fixed gelatin concentration. To further modify the bioink, a secondary, postprinting crosslinking step was used to adjust the modulus and degradation properties of the gel. Printed PEGX–gelatin was further crosslinked by exposure to N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDC) and N-hydroxysuccinimide (NHS) increasing the modulus from Pa to kPa. Normal degradation of the gel occurred after 2 days, but after secondary crosslinking degradation was extended to 4 weeks. Different bioinks were successfully coprinted and were capable of directing cell adhesion. Patterning of PEGX–PEG and PEGX–gelatin regulated human dermal fibroblasts to gelatin locations. Co-cultures of human umbilical vein endothelial cells (HUVECs) and human mesenchymal stem cells (hMSCs) were spatially organized in 3D by printing PEGX–gelatin seeded with HUVECs in a micro grid and filling the open spaces with hMSCs. After two weeks the gel had degraded but cells had formed a robust tissue with the grid pattern still observable. The customizable bioinks offer synthetic and natural bioinks for a variety of tissue engineering applications and further expands the number of bioinks available for 3D printing. The tunable material properties and degradation times show promise for future cell-specific bioinks.
To better represent the complexity of ECM in vivo, Pati et al.8 utilized decellularized extracellular matrix (dECM) for bioink formation. Unlike previously mentioned bioinks, the dECM does not require crosslinkers and can allow the cells contained within to degrade the surrounding gel. Multiple dECM bioinks were investigated for specific tissues, including adipose, cartilage, and heart tissues (adECM, cdECM, and hdECM respectively) (Fig. 2a). Decellularization of the ECM of each tissue required physical, chemical, and enzymatic processes with approximately 98% reduction of cellular contents of the tissues. The dECM was solubilized to a final concentration of 3% and adjusted to a physiological pH. The dECM pre-gel remained as a solution at temperatures below 15 °C and transformed to a gel after 30 min at 37 °C. 3D, porous structures of the dECMs were fabricated with a polycaprolactone (PCL) framework as seen in Fig. 2b. For each type of dECM a different strategy was used for scaffold design. The hdECM bioink was used to form a 3D structure without the use of PCL, but 200 μm lines of PCL were used for cdECM structures for increased load bearing support, and 100 μL lines of PCL were chosen for adECM structures for reduced stiffness. To examine cellular function within the dECM tissues, human adipose-derived stems cells (hASCs) and human inferior turbinate-tissue derived mesenchymal stromal cells (hTMSCs) were integrated into the adipose and cartilage dECMs respectively. Stem cell gene expression in the dECM gels was compared to cells cultured in collagen scaffolds. In the adipose scaffold, hASCs had an increase in the adipogenic marker lipoprotein lipase and hTMSCs demonstrated increased expression of the messenger RNA, SOX9, an early chondrogenic lineage factor. Results demonstrate cells cultured in the dECM scaffolds had increased the commitment of the stem cells to either adipogenic lineage or chondrogenic lineage. The functional maturation of rat myoblasts was used to verify the hdECM as a viable scaffold. A significant increase in cardiac-specific genes, fast myosin heavy chain and alpha-sarcomeric actinin, was observed when compared to myoblast genes in collagen frameworks. Overall, the 3D printed dECM bioinks supported differentiation and maturation of the three tissue specific cells. The dECM bioinks presented are an attractive option for in vitro and in vivo tissue development and an alternative to chemically crosslinked bioinks. The customized ECMs of the bioinks have applications in understanding complex cell–ECM interactions and tissue reconstruction.
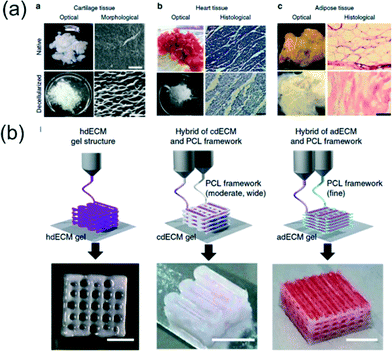 | ||
| Fig. 2 3D printing of dECM bioinks. (a) Optical and microscopic images of native and decellularized cartilage (scale bar, 50 μm), heart tissue (scale bar, 100 μm), and adipose tissue (scale bar, 100 μm). (b) Printing process of particular tissue constructs with dECM bioink. Heart tissue construct was printed only with heart dECM (hdECM). Cartilage and adipose tissues were printed with cartilage dECM (cdECM) and adipose dECM (adECM), respectively and in combination with a PCL framework (scale bar, 5 mm). Reprinted from ref. 8 with permission from Nature Publishing Group. | ||
Hydrogel scaffolds
Hydrogel templates and constructs are an ideal 3D scaffolding material as they have a range of cellular applications including cell storage, cell culture, monitoring cell–cell interactions, and cell actuation.9 They offer geometric control of the microenvironment allowing single-cell orientation to be guided and controlled. Unlike bioinks, 3D hydrogel scaffolds do not require extrusion, but can be bioprinted with templating methods and photoactivation. Hydrogel templates can constrict cells in tubular environments which have demonstrated the ability to replicate blood vessels.10 The dynamic range of hydrogel properties makes them advantageous for 3D tissue engineering and biomedical applications.For engineering hydrogel constructs for vascularization applications, Bertassoni et al.11 used 3D bioprinting of agarose fibers. Agarose gel was deposited through a dispensing capillary coordinated with an X–Y–Z stage. Hydrogel precursors, including methacrylated gelatin (GelMA), star poly(ethylene glycol-co-lactide) acrylate (SPELA), poly(ethyleneglycol) dimethacrylate (PEGDMA) and poly(ethylene glycol) diacrylate (PEGDA), were cast over the agarose fiber templates and photocrosslinked using UV light (Fig. 3a). The crosslinked hydrogels did not adhere to the agarose, allowing removal of the fibers with a light vacuum for microchannel formation within the gel (Fig. 3b). To mimic the dimensions of native blood vessels, microchannels with diameters ranging from 1000 μm to 150 μm were bioprinted in the hydrogels. Multiple channels of different dimensions and layers of channels were shown to be fabricated within the same gel scaffold. Cell viability of mouse calvarial pre-osteoblast cells (MC3T3) was investigated in GelMA hydrogels with microchannels and without (blocks). Cells embedded in the GelMA gel perfused with microchannels were analyzed with a live/dead assay and had ~90% viability compared to only 60% cell viability using the block hydrogel after 7 days of cell culture. Osteogenic differentiation of MC3T3s was also evaluated with an alkaline phosphatase (ALP) assay. The significant increased ALP activity of cells within the microchannel-GelMA compared to cells within the gel block indicated 3D microchannel gels promoted differentiation of the encapsulated cells. Cell culture within 250, 500, and 1000 μm diameter microchannels in GelMA was also accessed with HUVECs. Cells were able to form confluent endothelial monolayers within all three channel dimensions after 7 days of culture confirming the constructs remained perfusable and capable of vascularization (Fig. 3c–e). The proposed bioprinting technique has resulted in 3D hydrogels for increased cell viability and differentiation and customized microvascular networks for tissue engineering.
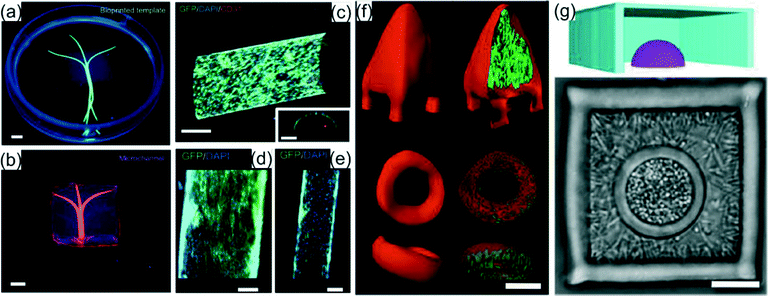 | ||
| Fig. 3 Examples of hydrogel scaffolding. (a) Photograph of the bioprinted templates (green) enclosed in GelMA hydrogels and (b) the respective microchannels perfused with fluorescent microbead suspension (pink). (c) View of z-stacked confocal images of HUVEC-lined microchannel and cross-sectional view (scale bars, 250 μm). (d, e) Proliferation of HUVECs in 1000 and 500 μm channels (scale bars, 250 μm). Reprinted from ref. 11 with permission from the Royal Society of Chemistry. (f) Confocal fluorescence isosurfaces show isolated P. aeruginosa microcolonies within a surface anchored 2 pL pyramid and an untethered 3 pL torus. (g) Cut-away 3D mask reconstruction (upper) and bright-field image (lower) depict examples of nested polymicrobial communities. S. aureus microclusters are confined in a hemispherical cavity surrounded by P. aeruginosa. Inner cavities are 1 pL and outer chambers are 30 pL. 5 μm thick roofs to seal the cavities are not visible. Reprinted from ref. 12 with permission from the National Academy of Sciences. | ||
Other than mammalian cells, 3D bioprinting has applications for bacteria cell culture. Using a direct laser writing approach based on multiphoton lithography (MPL), Connell et al.12 investigated the interaction of multiple bacteria colonies by engineering picoliter 3D culture environments. Bacteria were mixed in a warm solution composed of gelatin, bovine serum albumin, and photosensitive molecules (methylene blue or Rose Bengal) for crosslinking. The bacteria solution was allowed to cool to ambient temperatures to form a gel and suspend the bacteria. Enclosures of bacteria were fabricated by scanning a focused 740 nm titanium:sapphire laser in three dimensions within the gel to create walled structures with sub-micrometer resolution by crosslinking the BSA and gelatin (Fig. 3f). Although crosslinked, the laser fabricated barriers are permeable to cell culture nutrients allowing the bacteria within the structures, Pseudomonas aeruginosa and Staphylococcus aureus, to have doubling times of 45 and 35 minutes respectively at 37 °C. Bacteria were shown to be viable over multiple hours demonstrating the encapsulation process does not hinder cellular function. Co-culture environments of bacteria were fabricated by sequential printing of a micro spherical chamber of S. aureus surrounded by a cube of P. aeruginosa (Fig. 3g). Densities of initial cell populations could be varied to control the final cell density for each species. To investigate the cell interactions between the two bacteria, a S. aureus colony surrounded by a modified β-lactam resistant P. aeruginosa colony was exposed to β-lactam-based antibiotics. After exposure to antibiotics, the colony of S. aureus encapsulated with P. aeruginosa had a significantly higher survival rate (~80%) compared to S. aureus surrounded by wild type P. aeruginosa with no β-lactam resistance (~40%). The controlled nesting of bacteria colonies within hydrogels is a valuable tool for investigating cell–cell interactions, bacteria metabolic analysis, and understanding development of bacterial infections in vivo.
Conclusion and outlook
The recently published, highlighted works offer a glimpse into future 3D bioprinting techniques and scaffold design for biomedical applications. 3D printing of hydrogels offers structural organization with fine-tuned material properties including elastic modulus and chemical composition to control cell behavior. Bioink and hydrogel scaffolds offer artificial 3D environments comparable to in vivo tissue. The 3D constructs have proven to promote cell viability, differentiation, migration, and cell–cell interactions that are not observed in 2D culture systems. The future challenges for 3D bioprinting will include increased fabrication speed13 so scaffolds can be relevant for clinical applications and the design of bioinks and hydrogels that can be inexpensively produced for industrial biofabrication. Since one material or scaffold will not be versatile enough for multiple biomaterials, functionally adaptive gels that change shape or stiffness using biological stimuli will also need to be investigated. Materials science, cell biology, biophysics, and biomedical engineering will all be necessary to take on these challenges to move 3D bioprinting of hydrogels to real-world biomedical applications.Acknowledgements
Authors thank the European Research Council (ERC) for Starting Grant “Lab-in-a-tube and Nanorobotics biosensors; LT-NRBS” [no. 311529] for financial support.Notes and references
- L. Casares, R. Vincent, D. Zalvidea, N. Campillo, D. Navajas, M. Arroyo and X. Trepat, Nat. Mater., 2015, 14, 343–351 CrossRef CAS PubMed.
- E. Fennema, N. Rivron, J. Rouwkema, C. van Blitterswijk and J. de Boer, Trends Biotechnol., 2013, 31, 108–115 CrossRef CAS PubMed.
- W. Xi, C. K. Schmidt, S. Sanchez, D. H. Gracias, R. E. Carazo-Salas, S. P. Jackson and O. G. Schmidt, Nano Lett., 2014, 14, 4197–4204 CrossRef CAS PubMed.
- M. M. Stanton, J. M. Rankenberg, B.-W. Park, W. G. McGimpsey, C. Malcuit and C. R. Lambert, Macromol. Biosci., 2014, 14, 953–964 CrossRef CAS PubMed.
- S. V. Murphy and A. Atala, Nat. Biotechnol., 2014, 32, 773–785 CrossRef CAS PubMed.
- C. Li, A. Faulkner-Jones, A. R. Dun, J. Jin, P. Chen, Y. Xing, Z. Yang, Z. Li, W. Shu, D. Liu and R. R. Duncan, Angew. Chem., Int. Ed., 2015, 54, 3957–3961 CrossRef CAS PubMed.
- A. L. Rutz, K. E. Hyland, A. E. Jakus, W. R. Burghardt and R. N. Shah, Adv. Mater., 2015, 27, 1607–1614 CrossRef CAS PubMed.
- F. Pati, J. Jang, D.-H. Ha, S. Won Kim, J.-W. Rhie, J.-H. Shim, D.-H. Kim and D.-W. Cho, Nat. Commun., 2014, 5, 3935 CAS.
- L. Ionov, Adv. Funct. Mater., 2013, 23, 4555–4570 CrossRef CAS PubMed.
- R. Arayanarakool, A. K. Meyer, L. Helbig, S. Sanchez and O. G. Schmidt, Lab Chip, 2015 10.1039/C5LC00024F.
- L. E. Bertassoni, M. Cecconi, V. Manoharan, M. Nikkhah, J. Hjortnaes, A. L. Cristino, G. Barabaschi, D. Demarchi, M. R. Dokmeci, Y. Yang and A. Khademhosseini, Lab Chip, 2014, 14, 2202–2211 RSC.
- J. L. Connell, E. T. Ritschdorff, M. Whiteley and J. B. Shear, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 18380–18385 CrossRef CAS PubMed.
- J. R. Tumbleston, D. Shirvanyants, N. Ermoshkin, R. Janusziewicz, A. R. Johnson, D. Kelly, K. Chen, R. Pinschmidt, J. P. Rolland, A. Ermoshkin, E. T. Samulski and J. M. DeSimone, Science, 2015, 347, 1349–1352 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |
