Research highlights: aptamers on a chip
Donghyuk
Kim
,
Ivan
Pushkarsky
,
Andy
Tay
and
Dino
Di Carlo
*
Department of Bioengineering, California NanoSystems Institute, Jonsson Comprehensive Cancer Center, University of California Los Angeles, 420 Westwood Plaza, 5121 Engineering V, Box 951600, Los Angeles, California 90095, USA. E-mail: dicarlo@ucla.edu
First published on 24th February 2015
Abstract
Aptamers, oligonucleic acid or peptide molecules with binding affinity to a specific molecule, have gained broad scientific attention due to their stability, ease of production and modification, and durability, and therefore have been employed in a wide range of applications both in basic research and clinical science. Recent advances in high-throughput sequencing are poised to revolutionize the selection of aptamers, while microfluidic and microarray approaches have helped automate these processes of selection. Here we highlight work addressing several challenges in using aptamers more widely. We discuss array-based discovery of multivalent aptamers which is used to develop high affinity or paired aptamers by cleverly selecting new aptamers that bind to previously aptamer-bound proteins. Other highlighted work is addressing problems in analyzing local cell secretions, as well as refreshable sensors that detect signals over hundreds of cycles and can be refreshed with DI water scavenged from the air, leading to less reagent storage towards wearable sensors.
Array-based discovery of multivalent aptamers
Despite the impact of aptamers on the development of biosensors,1 a current bottleneck lies in the discovery of multiple aptamers that can recognize distinct non-overlapping epitopes on a common target.2 Multivalent receptor–ligand interactions can offer dramatically higher affinity and specificity when compared to monovalent counterparts.3 Additionally, non-overlapping binding epitopes are advantageous for sandwich sensing assays. However, the current aptamer discovery methods (i.e., systematic evolution of ligands by exponential enrichment (SELEX) as an example of the gold standard4) strongly tend to yield aptamers that only bind to the primary, highest affinity epitope.5 Furthermore, in current approaches binding measurements are performed serially for every combination of potential binding partners, which naturally leads to an extremely low throughput in the aptamer pair identification process.6To answer the current need of the field, Cho et al. developed a systematic screening method, that they call array-based discovery platform for multivalent aptamers (AD-MAP), to efficiently identify aptamer pairs. Using this AD-MAP, they discovered novel aptamer pairs for human angiopoietin-2 (Ang2) with the dissociation constant as low as 97 pM, which is an ~200-fold improvement compared to that of the individual component aptamers.7
They first performed four rounds of microfluidic SELEX with Ang2-coated beads, which resulted in about 30 million potential candidate aptamer sequences after high-throughput sequencing (HTS) (Fig. 1). Microfluidic SELEX automates sample preparation steps of the SELEX process to isolate and wash selection beads. Then, they selectively synthesized the 235 most-enriched sequences identified by HTS on an aptamer microarray. From this array, they fluorescently identified an aptamer sequence, termed as Ang2-binding aptamer 1 (ABA1), with the highest binding affinity (Kd = 20.5 ± 7.33 nM). Then, the ABA1–Ang2 complex and the aptamer array were used again to identify a set of aptamers that can still bind to the ABA1–Ang2 complex. Monitoring the signal difference between arrays challenged by ABA1–Ang2 and Ang2 alone, Cho et al. successfully identified three aptamers (ABA65, ABA92, and ABA109 with a Kd range of 37.6–57.0 nM). These three aptamers were examined to see if they could form a non-overlapping binding pair with ABA1 both in standard buffer conditions and in a sample biological matrix (undiluted fetal bovine serum (FBS)) using an enzyme-linked oligonucleotide assay (ELONA) where they observed ~20 nM limit of detection for Ang2 in undiluted FBS. Upon demonstration of the above aptamers, Cho et al. synthesized a physically linked aptamer pair (ABA1–ABA65, a combination with the highest and second-highest affinity aptamers). For linking, they used a flexible poly-T linker with different lengths to examine potential interference from the linker. Among the 5, 10, 16, and 25 poly-T linkers (5T, 10T, 16T, and 25T), the 25T-linked ABA1–ABA65 pair showed the greatest increase in binding affinity using a bead-based fluorescence assay. The Kd for the ABA1–ABA65 pair was observed to be 97 pM which is ~210-fold higher than ABA1 alone or ~390-fold higher than ABA65 alone, successfully demonstrating the concept of bidentate binding and proving the power of their technique to identify aptamer pairs. The 25T linker alone, or ABA1 and ABA65 each of which conjugated to only the 25T linker, did not show such an increase in affinity, indicating that such an increase was not due to the linker.
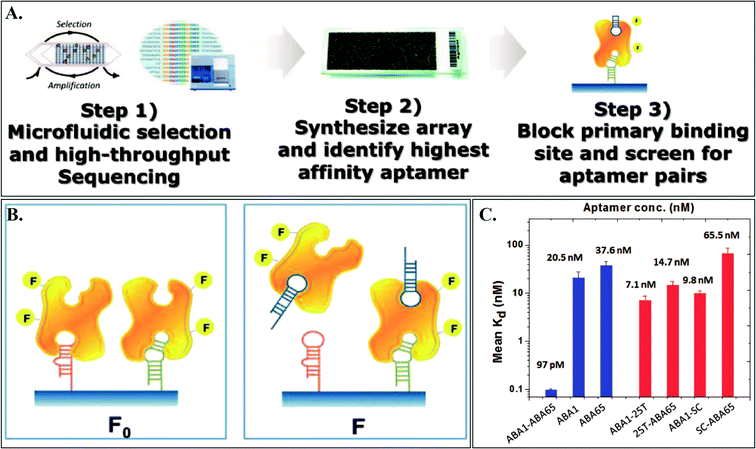 | ||
| Fig. 1 A. Overview of the AD-MAP process where a pool of aptamer candidates are obtained by microfluidic SELEX (step 1), the sequences with the highest copy number are synthesized on a custom array (step 2), and aptamer pairs that can bind to different epitopes on the target simultaneously are screened on the array (step 3). B. Fluorescence measurement to identify aptamer pairs: F0 – fluorescence from the array challenged with Ang2, F – fluorescence from the array challenged with ABA1–Ang2. C. Binding assays with various controls (individual component and linkers) demonstrating that the linked aptamers ABA1 and ABA65 contribute significantly to the enhanced affinity. Figures adapted from the work by Cho et al.7 with permission. | ||
In summary, Cho et al. demonstrated an approach for systematic discovery of aptamer pairs that are capable of simultaneous binding to two different binding sites on a target protein. The use of aptamer arrays made the selection and sequencing process highly parallel and thus increased the throughput of the entire process significantly, which in turn may enable the development of more sensitive diagnostic tools or improve the targeting effectiveness of aptamer-based drugs.
Aptamers for electrochemical sensing of TGF-β1 release from liver cells
Liver fibrosis is characterized by the build-up of excessive extracellular matrix (ECM) in the liver as a result of inflammation caused by liver injury. This build-up of ECM leads to the hardening of liver tissue which diminishes liver tissue function and advanced liver fibrosis may lead to cirrhosis or liver failure which often requires a liver transplant. Specifically, hepatic stellate cells (HSCs), which are normally in a quiescent state become activated following an injury to the liver, causing them to secrete the fibrogenic cytokine transforming growth factor beta 1 (TGF-β1). TGF-β1, in turn, triggers other HSCs to assume a more contractile myofibroblast-like phenotype and exert traction forces on local tissues causing an increase in their stiffness. The increased stiffness then stabilizes this activated myofibroblast-like phenotype leading to further ECM deposition as well as secretion of TGF-β1, perpetuating this positive feedback loop. Fortunately, liver fibrosis has been shown to be potentially reversible at early stages, mainly through the inactivation of the HSCs.8 It is therefore important to understand how fast fibrotic triggers such as TGF-β1 appear, and what their dynamics are post-injury. Towards this end, Matharu et al. have developed a microfluidic platform that uses aptasensors to electrochemically monitor the dynamics of TGF-β1 released by activated HSCs over 20 hours.9In principle, the electrochemical sensor works by recording the change in the efficiency of electron transfer from a redox reporter (coupled to a molecule) to a reference electrode. If a target-specific sensing molecule is coupled to a redox reporter on one end and to an electrode on the other end, the efficiency of electron transfer can be modulated by the configuration or folding of this molecule due to it binding its target. This kind of biosensor has been extensively described previously.10 In this work, the sensing molecule used was a TGF-β1-specific aptamer. This aptamer was coupled to a redox reporter (Methylene Blue) and immobilized onto an array of gold electrodes through self-assembly using Au–thiol chemistry, within a microfluidic device. The aptamers would be able to bind TGF-β1 released by nearby seeded hepatic stellate cells, causing a change in their configuration and, therefore, the measured redox current.
The authors performed several validation steps for this assay. First, modification of the Au surface with aptamers and the binding of TGF-β1 to the aptamer were characterized using surface plasmon resonance (SPR) and taking the SPR angle difference between the initial and modified surface. Next, the sensitivity of the aptamer was determined using square wave voltammetry experiments with a modified Au electrode challenged with varying concentrations of TGF-β1. Finally, specificity of the aptamer was tested by performing the same experiment but challenging the chip with other proteins such as immunoglobulin G (IgG), bovine serum albumin (BSA), interleukin-2 (IL-2) and others. These non-specific proteins were found to yield only 10% of the signal given by TGF-β1.
In order to prevent cells from adhering to the electrodes and potentially causing noise in the readout, the authors devised a reconfigurable microfluidics device to protect the electrodes during the seeding step consisting of two PDMS layers mounted on a glass slide with micro-patterned Au electrodes (Fig. 2A). The middle polydimethylsiloxane (PDMS) layer is the working layer with micro-channels for seeding cells. Each of these microchannels contains four micro-cups (whose heights are shorter than those of the microchannels) that are aligned directly above the Au electrodes on the glass slide. The top PDMS layer is the control layer consisting of one large channel which, when pressurized, presses the microcups into contact with the glass slide below, shielding the Au electrodes from the rest of the microchannel (Fig. 2B).
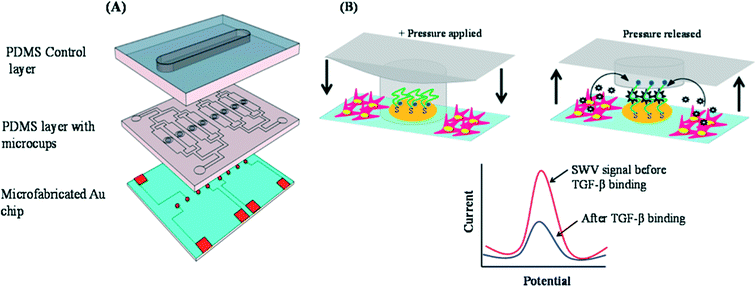 | ||
| Fig. 2 (A) The three layers of the microfluidic device. Bottom: glass slide with micropatterned Au electrodes. Middle: working PDMS layer with fluidic microchannels and microcups. Top: pressurizable control PDMS layer for controlling the microcups. (B) Diagram showing the actuation of the microcups. The microcups are lowered during the protein adsorption and cell-seeding phases, then raised for the sensing phase. The plot gives a general description of the sensing mechanism. As TGF-β1 is bound by the aptamer, electron transfer efficiency between the redox reporter and electrode is reduced. Figures adapted from the work by Matharu et al.9 with permission. | ||
To demonstrate the platform's performance, collagen was adsorbed to the glass slide and LX2 cells (a human hepatic stellate cell line) were seeded and allowed to adhere and spread while the electrodes were shielded by the microcups. The microcups were then lifted and the cells were activated by infusion of platelet-derived growth factor (PDGF). The cells were cultured for 20 h and a comparison of TGF-β1 levels between PDGF-activated and quiescent cells revealed a 6-fold increase of TGF-β1 levels in the PDGF-activated cells.
While secretion and the levels of TGF-β1 are clearly important, the contractile phenotype of the hepatic stellate cells could also be characterized in such a platform leading to links between molecular and disease-causing phenotypic characteristics of these cells that will be important to understand to treat liver fibrosis.
Maintenance-free aptamer biosensor
The use of aptamers in long-term monitoring applications, such as in wearable devices or in a remote environment, has been limited by low off rates and subsequent slowly reversible sensor responses. Repetitive detection often requires removal of analyte-bound aptamers and introduction of fresh ones, or wash solutions that denature aptamer structure followed by refreshing solutions. Both of these approaches can be difficult to implement in an unattended continuous monitoring system. In their paper, Potyrailo et al. created a thrombin aptamer-based biosensor that outperformed all previously described biosensors in two aspects: (1) aptamer reversibility over larger number of bind–release cycles; and (2) potential for a maintenance-free system.11 Their design is capable of up to 365 reversible bind–release cycles and can be reversibly deactivated with scavenged deionized (DI) water up to 100 times.The group first conjugated biotin-linked oligonucleotides of the thrombin aptamers to a streptavidin-functionalized chip. Next, buffer was introduced to fold the aptamer into their target-binding conformation. After target binding, the aptamers were deactivated using either sodium hydroxide–sodium chloride (NaOH–NaCl) or DI water (Fig. 3a).
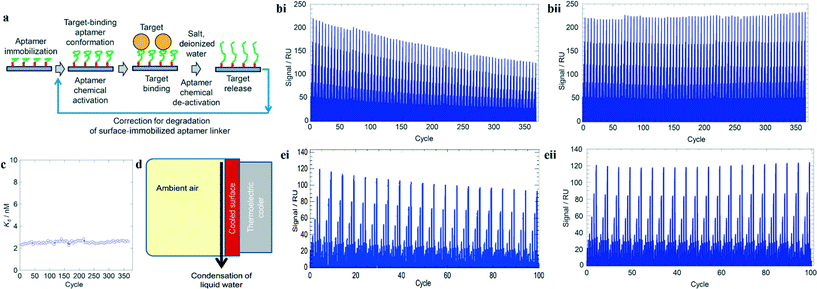 | ||
| Fig. 3 Aptamer-based biosensor operating with high number of reversible bind–release cycles. (a) Steps to construct aptamer-based sensor. Deactivating reagents can be salt or DI water. (bi) Reduction of 40% signal after 365 bind–release cycles with NaOH–NaCl. (bii) Signal after correction for streptavidin–biotin dissociation. (c) Stable binding affinity (Kd) of immobilized aptamers over 365 bind–release cycles. (d) Conceptual design for scavenging water from ambient environment. (ei) Reduction of 20% signal after 100 bind–release cycles with DI water. (eii) Signal after correction for streptavidin–biotin dissociation. Figures adapted from the work by Potyrailo et al.11 with permission. | ||
Interestingly, it was found that the signal from surface plasmon resonance showed a 40% reduction over 365 bind–release cycles (Fig. 3bi) although the binding affinity (Kd) of the aptamer for thrombin remained stable (Fig. 3c). This motivated the investigation of the streptavidin–biotin linkage which was found to have a dissociation rate 1.5–3 times faster than previously determined. After factoring in this correction to their data, the new signal showed the high stability of the binding aptamers over many bind–release cycles (Fig. 3bii).
The authors also explored the idea of a maintenance-free biosensor by utilizing a thermoelectric cooler to scavenge DI water from ambient air for deactivating the aptamers (Fig. 3d). Similarly, the 20% reduction in signals was corrected after accounting for the dissociation between the streptavidin and biotin (Fig. 3ei–ii). However, the use of DI water as a deactivating reagent doubled the time for each bind–release cycle and the reversible operations were only performed up to 100 bind–release cycles due to the lack of salts. Furthermore, while the DI water may be obtained freely from the environment, the scavenged volume depends on the humidity and a reservoir of activating buffer is still needed to automate biosensing.
In conclusion, the aptamer biosensor described here demonstrates the potential of performing multiple bind–release cycles without significant loss of specificity of the aptamers. Also, the authors highlighted the importance of characterizing the immobilization molecules used as the loss in signal may not always be due to degradation of aptamers or bioreceptors. Nonetheless, the sensitivity loss due to streptavidin–biotin dissociation can become an issue for an unattended sensor that requires more than one sampling a day or a sensor that requires deactivating reagents with high salt content. Hence, more research is warranted to truly develop a maintenance-free sensor.
References
- B. E. Collins and J. C. Paulson, Curr. Opin. Chem. Biol., 2004, 8, 617–625 CrossRef CAS PubMed.
- F. Liu and K. J. Walters, Trends Biochem. Sci., 2010, 35, 352–360 CrossRef CAS PubMed.
- D. Neri, M. Momo, T. Prospero and G. Winter, J. Mol. Biol., 1995, 246, 367–373 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CAS.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- S. Fitter and R. James, J. Biol. Chem., 2005, 280, 34193–34201 CrossRef CAS PubMed.
- M. Cho, S. S. Oh, J. Nie, R. Stewart, M. J. Radeke, M. Eisenstein, P. J. Coffey, J. A. Thomson and H. T. Soh, Anal. Chem., 2015, 87, 821–828 CrossRef CAS PubMed.
- X. Liu, J. Xu, D. A. Brenner and T. Kisseleva, Reversibility of Liver Fibrosis and Inactivation of Fibrogenic Myofibroblasts, Curr. Pathobiol. Rep., 2013, 1, 209–214 CrossRef PubMed.
- Z. Matharu, et al., Detecting Transforming Growth Factor-β Release from Liver Cells Using an Aptasensor Integrated with Microfluidics, Anal. Chem., 2014, 86, 8865–8872 CrossRef CAS PubMed.
- Y. Xiao, R. Y. Lai and K. W. Plaxco, Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing, Nat. Protoc., 2007, 2, 2875–2880 CrossRef CAS PubMed.
- R. A. Potyrailo, A. J. Murray, N. Nagraj, A. D. Pris, J. M. Ashe and M. Todorovic, Towards maintenance-free biosensors for hundreds of bind/release cycles, Angew. Chem., Int. Ed., 2015, 54(7), 2174–2178, DOI:10.1002/anie.201411094.
| This journal is © The Royal Society of Chemistry 2015 |
