Accurate microfluidic sorting of droplets at 30 kHz†
Adam
Sciambi‡
and
Adam R.
Abate
*
Department of Bioengineering and Therapeutic Sciences, California Institute for Quantitative Biosciences, University of California, San Francisco, California, 94158 USA. E-mail: adam.abate@ucsf.edu; Fax: +1 415 514 1028; Tel: +1 415 476 9819
First published on 22nd October 2014
Abstract
Fluorescence-activated droplet sorting is an important tool for droplet microfluidic workflows, but published approaches are unable to surpass throughputs of a few kilohertz. We present a new geometry that replaces the hard divider separating the outlets with a gapped divider, allowing sorting over ten times faster.
Droplet microfluidics,1 with its ability to compartmentalize reactions in sub-nanoliter volumes, enables hundreds of millions of distinct biological assays to be performed on individual biomolecules or cells. When performing so many distinct assays, it is often necessary to select for a subset of reactions for additional manipulation by means of droplet sorting.2 Droplet sorting has been used, for example, to enhance the activities of horseradish peroxidase3 and betagalactosidase,4 to screen mutant libraries for xylose consumption,5 and to enrich cancer cells out of a mixed population using single-cell PCR.6
The ability of the droplet sorter to detect rare events depends on its sorting speed, since fast sorters can screen large numbers of droplets. For this reason, there has been a sustained effort to develop faster droplet sorters, but existing systems are still much slower than commercially available fluorescence-activated cell sorters (FACS), which achieve sorting rates up to 50 kHz. For example, dielectrophoretic droplet sorters apply a brief electric field to deflect target droplets into a collection channel and achieve just 2 kHz.3,6–10 Selective coalescence sorters also achieve just 2 kHz,4,11 while surface acoustic wave devices achieve 3 kHz.12–14 In these devices, the factor that limits faster sorting is the use of a hard divider between the collection and waste channels. As flow rates increase to sort faster, the effects from viscosity become comparable to those from drop surface tension, and drops split at the divider rather than deflect intact. The transition occurs when the capillary number, Ca = vμ/γ, nears 1, where v is the characteristic velocity, μ is the dynamic viscosity, and γ is the interfacial tension. Empirically, above Ca ~ 0.1, drops split into both channels and contaminate the collection reservoirs. To reach faster sorting rates, new architectures are needed that overcome the tendency of droplets to split at the very high flow rates and capillary numbers required.
In this paper, we describe a microfluidic design that permits 30 kHz droplet sorting with >99% accuracy. This tenfold rate increase compared to the fastest available droplet sorters enables ~108 droplets to be sorted per hour and over a billion per day. Indeed, with our architecture, sorting speed is not limited by the physical mechanism of sorting (even at Ca ~ 1) but rather by the electronics that detect the droplets; with faster electronics, we anticipate even faster sorting.
The devices were fabricated using soft lithography of poly(dimethylsiloxane) (PDMS) moulded from device masters. The masters were created from two sequential layers (11 μm and 19 μm thick) of photoresist (MicroChem, SU-8 3010) spun onto a silicon wafer. Uncured PDMS consisting of a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polymer to cross-linker mixture (Dow Corning, Sylgard 184) was poured onto the master, degassed, and baked at 85 °C for 2 hours. The PDMS mould was then cut and peeled from the master, punched with a 0.75 mm Harris Uni-core for inlet ports, and plasma bonded to a 1 mm thick, 10
1 polymer to cross-linker mixture (Dow Corning, Sylgard 184) was poured onto the master, degassed, and baked at 85 °C for 2 hours. The PDMS mould was then cut and peeled from the master, punched with a 0.75 mm Harris Uni-core for inlet ports, and plasma bonded to a 1 mm thick, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PDMS slab to ensure a strong bond. The bonded PDMS device was then baked at 85 °C for 10 min. The bottom of the all-PDMS device was then plasma bonded to a glass slide to provide structural support and rigidity. To enable immediate usage of the device with water-in-oil emulsions, we performed a hydrophobic surface treatment by flushing with Aquapel, clearing with pressurized air and baking at 85 °C for an additional 30 min.
1 PDMS slab to ensure a strong bond. The bonded PDMS device was then baked at 85 °C for 10 min. The bottom of the all-PDMS device was then plasma bonded to a glass slide to provide structural support and rigidity. To enable immediate usage of the device with water-in-oil emulsions, we performed a hydrophobic surface treatment by flushing with Aquapel, clearing with pressurized air and baking at 85 °C for an additional 30 min.
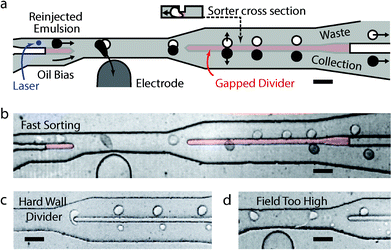
The primary innovation that allows us to increase sorting speed by over an order of magnitude is to replace the impermeable wall that usually divides the collection and waste channels with a gapped divider. The gapped divider, which reaches only part way from the channel ceiling to floor, allows droplets to squeeze into an energetically unfavourable region (11 μm tall) between the sort channels (30 μm tall). Due to the droplet Laplace pressure, small lateral displacements above or below the sorter centre line grow as the droplets travel downstream, pushing them fully into the nearest channel. The process is shown in the schematic of Fig. 1a, with a cross section of the squeezed drop in the gapped divider (inset). It is also depicted in the still in Fig. 1b taken from a high speed movie (see ESI†) of 25 μm droplets sorted at 22 kHz. This is ten-fold faster than conventional sorters, which use hard wall dividers that split droplets at similar flow rates due to shear at the divider edge (Fig. 1c). Splitting does not occur if droplets are displaced sufficiently beyond the divider before they reach its starting edge; however, at high flow rates, the large electric fields required break the droplets apart (Fig. 1d). By contrast, when the gapped divider is used, the droplets experience less shear and are able to gradually enter one channel intact.
Another factor critical for achieving maximum sorting speed is the minimization of the oil spacer flow rate. Proper droplet spacing is essential in allowing the droplets to be interrogated and sorted individually, but too much oil increases capillary number and limits sorting speed. To minimize oil flow rate, we used a narrow, 50 μm-wide sorting junction with a wide electrode, which exerted a constant force on the droplets over a long distance. We also implemented a second gapped divider at the entrance of the sorting junction (red section at the left of Fig. 1a and b), which pinned incoming droplets against the upper wall so that the full channel was used during sorting and no oil was wasted. This divider operated by diverting high flow-rate carrier oil, required to properly space droplets, from the reinjection channel into a lower channel carrying the bias oil used to tune lateral drop position downstream. The combined oil flow from below pinned incoming drops to the upper channel wall; without such a design, the droplets move to the centre of the channel.
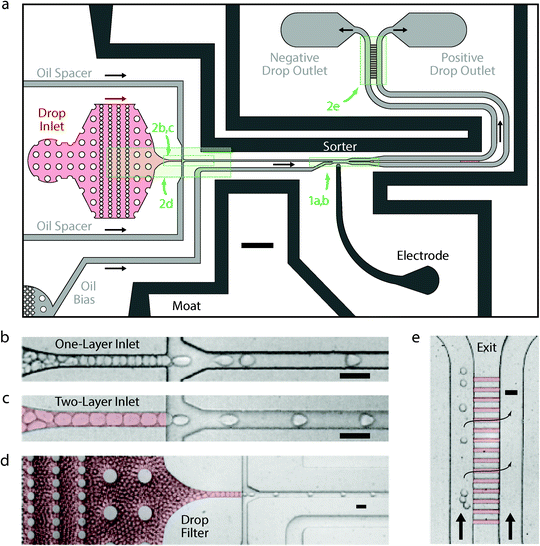
The gapped dividers allow us to maximize sorting speed, but other features are needed to ensure sorting accuracy (Fig. 2a). For example, as flow rates increase to sort faster, inlet pressures grow causing the droplet filter to bow. Bowing widens the filter gaps permitting dust to pass that clogs the device. It also causes droplets to pack in vertical layers, leading to irregular spacing (Fig. 2b) and sorting errors. To address these issues, we used an alternate design with the filter in the same shallow layer as the gapped divider (red drop inlet in Fig. 2a) rather than the taller layer of the rest of the sorting junction (coloured grey). The filter still bowed under the pressure, but the gaps remained small enough to remove debris. Moreover, as the droplets approached the injection channel, they were forced into a monolayered, single-file line for even spacing (Fig. 2c). Evidence of bowing can be seen in the open areas of the filter, where deformation around the posts appeared as non-uniform shading and where droplets stacked vertically in multiple layers (Fig. 2d).
After spacing, the droplets travelled to the sorter, as shown in Fig. 2a and expanded in Fig. 1a and b. There, the droplets were scanned by a laser and their fluorescence measured. A salt water electrode (2 M NaCl)15,16 connected to a high voltage amplifier applied the electric field that sorted the droplets. The moat, a grounded salt water electrode bordering the device, generated the field gradient necessary for dielectrophoretic deflection and limited stray fields that could cause unintended droplet merger in the filter. Once sorted, the divided populations travelled down two parallel channels with similar hydrodynamic resistance. Because the negatively sorted population was often much larger than the positively sorted, the negative channel experienced greater flow resistance from droplet drag. To equilibrate pressures and enable controlled dispensing into the collection reservoirs, a shallow series of parallel channels was included near the outlet (magnified in Fig. 2e) to allow oil, but not droplets, to move between outlets and equilibrate small pressure differentials. This also made the sorter less sensitive to small differences in the outlet tube heights, which could generate a gravitational back-pressure that could interfere with the droplet sorting.
To demonstrate the effectiveness of the fast sorter, we used it to sort a test emulsion consisting of two droplet populations: a dim “negative” population and a bright “positive” population. The positive droplets consisted of phosphate-buffered saline with 1.4% by volume 0.5 μm latex beads (Sigma Aldrich, L3280) to make them appear dark in the optical microscopy images (Fig. 1b) and 0.75% fluorescent yellow 0.03 μm latex beads (Sigma Aldrich, L5150) to make them brightly fluorescent. The negative droplets were phosphate-buffered saline with 0.13% fluorescent yellow beads, making them dimly fluorescent so that they too could be detected by our drop detector. To create sufficient emulsion for several hours of sorting at 30 kHz, the emulsions were generated using serial droplet splitting,17 formed initially as 50 μm droplets that were each halved three times to produce 8 droplets 25 μm in diameter. This enabled the generation of droplets at 2 mL h−1 for the aqueous phase, approximately five times faster than could be achieved with a flow focusing generator.
To sort the emulsion, the droplets were injected into the device at 0.7 mL h−1, with the drop spacing oil and drop position-tuning bias oil each at 7 mL h−1. That corresponded to an average flow velocity of 3 m s−1 through the 30 μm × 50 μm sorter cross section. The fluorinated oil (3 M, HFE-7500) and 1% PEG-PFPE amphiphilic block copolymer surfactant18 combined for a drop interfacial tension of 4 mN m−1 (ref. 19) and a nearly matched water–oil dynamic viscosity of 0.1 mPa s, giving a very large Ca of 0.8 at that flow. The fluorescence was generated by a 473 nm laser (CNI Lasers), filtered at 517 ± 10 nm by a bandpass filer (Semrock), and measured by a photomultiplier tube (PMT, Thorlabs, PMM02). The signal was analysed by an FPGA (NI, PCI-7833R) with custom LabVIEW software. Droplets falling within the user-defined thresholds were sorted via an amplified pulse (Trek 609E-6) from the FPGA, transmitted into the device via a salt water electrode.15 To visualize the sorting and capture high speed videos, the device was illuminated with infrared light that did not overlap spectrally with the droplet fluorescence and imaged with a fast camera (Phantom, Miro M310) at a 50 kHz frame rate.
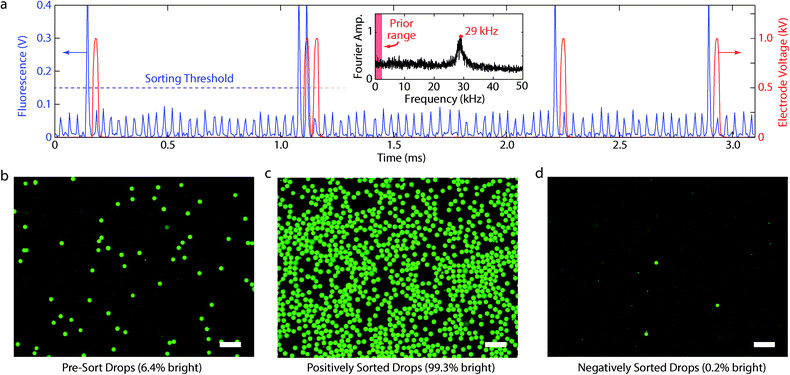
The fluorescence signals and pre- and post-sorted droplet populations from an hour-long, ~30 kHz sorting run (equivalent to processing over 108 drops) are shown in Fig. 3. As the droplets passed through the excitation laser, their emitted fluorescence was detected by the PMT, which outputted a voltage proportional to the intensity of the emitted light. The semi-periodic drop fluorescences, as detected by the PMT, are shown in a time series in blue in Fig. 3a, with the corresponding sorting pulses in red. The PMT had a bandwidth of 0–20 kHz, such that frequency components above this range were attenuated by a factor proportional to their fold-increase over 20 kHz. PMTs with higher frequency response are commercially available and can be implemented to detect droplets more quickly. The PMT voltages were recorded at 200 kHz, a sampling period of 5 μs, which was approximately the time a droplet spends in the detector region. The individual droplet signals, despite being broadened in time and attenuated in amplitude by the limited PMT bandwidth, were nevertheless still well above the noise floor and distinguishable as shown in the time trace.
Positive droplets were identified as those whose fluorescence was above a PMT threshold of 0.15 V and whose temporal width at the threshold was <50 μs, which excluded large, merged droplets. When a positive droplet was detected, the computer outputted a 1 V, 33 μs rectangular pulse amplified with edge rounding to 1 kV by the 13 kHz-bandwidth, high voltage amplifier. The detection laser was positioned ahead of the electrode so that, after identifying a droplet, an immediately-applied pulse corresponded to the moment the droplet was directly opposite the electrode, which was optimal for sorting. To estimate sorting speed, we measured the droplet rate in the time series of an hour long run by identifying peaks and, additionally, by measuring the maximum of the Fourier transform, which was centred on 29 ± 1 kHz (Fig. 3a, inset). The sorting rates of conventional microfluidic devices are ten-fold less than this and, for comparison, fall within the red band in the left of the inset.
To confirm accurate droplet sorting, we imaged the pre- (Fig. 3b), positive- (Fig. 3c), and negative-sort (Fig. 3d) droplet populations with fluorescence microscopy. The pre-sort population was 6.4% bright, in agreement with the fraction of positives detected with the PMT time traces. The positive population was 99.3% and the negative 0.2% bright. The false positives (dim droplets in the positive population) were abnormally large (most were >3 times the mean droplet volume, Fig. 3c) and were likely merged drops that were too large for the device design. The false negatives (bright droplets in negative population) were abnormally small (<2 the mean volume), which likely led to a proportionally smaller dielectrophoretic force and inadequate deflection during sorting. In most cases, sorting errors thus resulted from polydispersity in the starting emulsion, suggesting that higher accuracy requires more uniform emulsions. This is difficult to achieve because most emulsions, no matter the care taken to generate and handle them, will contain rare instances of droplets that merged or split and are thus abnormally large or small. Filtration of the emulsion prior to sorting may improve this, but requires additional steps that can result in even more merger and splitting.
Our device achieved sorting rates that rival those of fluorescence-activated cell sorters, which can sort at tens of kilohertz.20 Recently, small microfluidic droplets (10–20 μm) have been sorted at 10–15 kHz using these FACS methods.21,22 However, this required a double emulsification step in which the water-in-oil droplets were suspended as water-in-oil-in-water double emulsions in an aqueous carrier compatible with FACS. This may not be appropriate for all applications since double emulsions are generally less stable than single emulsions and, in addition, tend to be more permeable to small molecules, which can leach out of the droplets over time. In instances in which these issues are important, fast microfluidic droplet sorting is valuable.
Conclusions
We have presented a microfluidic device that accurately sorts droplets at 30 kHz, ten times faster than existing droplet sorters. Pushing the rate higher is possible but will require faster electronics. The speed of our droplet sorter will allow sorting of emulsions with unprecedented numbers of droplets. This will be valuable for applications in protein engineering and cell biology, in which the target droplets or cells are extremely rare in the population. Such enrichment is critical, for example, for enhancing enzymes through droplet-based microfluidic directed evolution or for isolating very rare circulating tumour cells from blood cell.Acknowledgements
This work was supported by an NSF CAREER Award (DBI-1253293), a grant from the NIH (HG007233-01), a Research Award from the California Institute for Quantitative Biosciences (QB3), the Bridging the Gap Award from the Rogers Family Foundation, a New Frontiers Research Award from the UCSF/Sandler Foundation Program for Breakthrough Biomedical Research, and a grant from the University of California Proof of Concept Program.Notes and references
- S.-Y. Teh, R. Lin, L.-H. Hung and A. P. Lee, Lab Chip, 2008, 8, 198–220 RSC.
- M. T. Guo, A. Rotem, J. A. Heyman and D. A. Weitz, Lab Chip, 2012, 12, 2146–2155 RSC.
- J. J. Agresti, E. Antipov, A. R. Abate, H. Ahn, A. C. Rowat, J.-C. Baret, M. Marquez, A. M. Klibanov, A. D. Griffiths and D. A. Weitz, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4004–4009 CrossRef CAS PubMed.
- A. Fallah-Araghi, J.-C. Baret, M. Ryckelynck and A. D. Griffiths, Lab Chip, 2012, 12, 882–891 RSC.
- B. J. Hindson, K. D. Ness, D. A. Masquelier, P. Belgrader, N. J. Heredia, A. J. Makarewicz, I. J. Bright, M. Y. Lucero, A. L. Hiddessen, T. C. Legler, T. K. Kitano, M. R. Hodel, J. F. Petersen, P. W. Wyatt, E. R. Steenblock, P. H. Shah, L. J. Bousse, C. B. Troup, J. C. Mellen, D. K. Wittmann, N. G. Erndt, T. H. Cauley, R. T. Koehler, A. P. So, S. Dube, K. A. Rose, L. Montesclaros, S. Wang, D. P. Stumbo, S. P. Hodges, S. Romine, F. P. Milanovich, H. E. White, J. F. Regan, G. A. Karlin-Neumann, C. M. Hindson, S. Saxonov and B. W. Colston, Anal. Chem., 2011, 83, 8604–8610 CrossRef CAS PubMed.
- D. J. Eastburn, A. Sciambi and A. R. Abate, Nucleic Acids Res., 2014, 42, e128 CrossRef PubMed.
- K. Ahn, J. Agresti, H. Chong, M. Marquez and D. A. Weitz, Appl. Phys. Lett., 2006, 88, 264105 CrossRef PubMed.
- L. Mazutis, J. Gilbert, W. L. Ung, D. A. Weitz, A. D. Griffiths and J. A. Heyman, Nat. Protoc., 2013, 8, 870–891 CrossRef CAS PubMed.
- J.-C. Baret, O. J. Miller, V. Taly, M. Ryckelynck, A. El-Harrak, L. Frenz, C. Rick, M. L. Samuels, J. B. Hutchison, J. J. Agresti, D. R. Link, D. A. Weitz and A. D. Griffiths, Lab Chip, 2009, 9, 1850–1858 RSC.
- B. L. Wang, A. Ghaderi, H. Zhou, J. Agresti, D. A. Weitz, G. R. Fink and G. Stephanopoulos, Nat. Biotechnol., 2014, 32, 473–478 CrossRef CAS PubMed.
- L. M. Fidalgo, G. Whyte, D. Bratton, C. F. Kaminski, C. Abell and W. T. S. Huck, Angew. Chem., Int. Ed., 2008, 47, 2042–2045 CrossRef CAS PubMed.
- T. Franke, S. Braunmüller, L. Schmid, A. Wixforth and D. A. Weitz, Lab Chip, 2010, 10, 789–794 RSC.
- X. Ding, S.-C. S. Lin, M. I. Lapsley, S. Li, X. Guo, C. Y. Chan, I.-K. Chiang, L. Wang, J. P. McCoy and T. J. Huang, Lab Chip, 2012, 12, 4228–4231 RSC.
- L. Schmid, D. A. Weitz and T. Franke, Lab Chip, 2014, 14, 3710–3718 RSC.
- A. Sciambi and A. R. Abate, Lab Chip, 2014, 14, 2605–2609 RSC.
- D. Eastburn, A. Sciambi and A. Abate, Anal. Chem., 2013, 85, 8016–8021 CrossRef CAS PubMed.
- A. R. Abate and D. A. Weitz, Lab Chip, 2011, 11, 1911–1915 RSC.
- C. Holtze, R. A. C, J. J. Agresti, J. B. Hutchison, F. E. Angilè, C. H. J. Schmitz, S. Köster, H. Duan, K. J. Humphry, R. A. Scanga, J. S. Johnson, D. Pisignano and D. A. Weitz, Lab Chip, 2008, 8, 1632–1639 RSC.
- A. R. Abate, P. Mary, V. van Steijn and D. A. Weitz, Lab Chip, 2012, 12, 1516–1521 RSC.
- A. Aharoni, K. Thieme and C. Chiu, Nat. Methods, 2006, 3, 609–614 CrossRef CAS PubMed.
- S. W. Lim and A. R. Abate, Lab Chip, 2013, 13, 4563–4572 RSC.
- A. Zinchenko, S. R. A. Devenish, B. Kintses, P.-Y. Colin, M. Fischlechner and F. Hollfelder, Anal. Chem., 2014, 86, 2526–2533 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4lc01194e |
| ‡ Current address: Mission Bio, Inc., San Francisco, California, 94107 USA. |
| This journal is © The Royal Society of Chemistry 2015 |