Enhanced propionic acid production from whey lactose with immobilized Propionibacterium acidipropionici and the role of trehalose synthesis in acid tolerance†
Ling
Jiang
a,
Huaiyan
Cui
b,
Liying
Zhu
c,
Yi
Hu
b,
Xian
Xu
b,
Shuang
Li
b and
He
Huang
*b
aCollege of Food Science and Light Industry, Nanjing Tech University, No. 5 Xinmofan Road, Nanjing 210009, People's Republic of China
bCollege of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, No. 5 Xinmofan Road, Nanjing 210009, People's Republic of China. E-mail: biotech@njtech.edu.cn; Fax: +86 25 58139942
cCollege of Sciences, Nanjing Tech University, No. 5 Xinmofan Road, Nanjing 210009, People's Republic of China
First published on 13th August 2014
Abstract
Whey lactose is a major by-product of the cheese and casein manufacturing industry. Utilization of this cheap and renewable resource is an area of interest among biotechnologists and bulk chemists. The objective of the present work was to determine the suitability of whey lactose for propionic acid production by a food microbe, Propionibacterium acidipropionici. Batch and fed-batch fermentations of whey lactose with both free cells and immobilized cells in a fibrous bed bioreactor (FBB) were examined and compared. It was observed that cells immobilized on polyethylenimine-treated Poraver (PEI-Poraver) in FBB favored higher productivity and yield of propionic acid as compared to free-cell fermentation. Interestingly, P. acidipropionici accumulated high levels of trehalose, especially in response to acid stress. An analysis of the P. acidipropionici genome sequence revealed the presence of two putative trehalose biosynthesis pathways (OtsA–OtsB and TreY–TreZ), and their functions in response to acid stress were subsequently studied. To further improve propionic acid production, an enhanced trehalose synthesis mutant was obtained by over-expression of an otsA gene encoding enzyme belonging to the OtsA–OtsB pathway. In this mutant, the maximum concentration of propionic acid reached 135 ± 6.5 g L−1 in a fed-batch fermentation approach, which to our knowledge is among the highest propionic acid concentration ever produced in the traditional fermentation process. In view of these results it was concluded that the FBB fed-batch fermentation with an enhanced trehalose synthesis mutant from whey lactose can therefore be considered as a good candidate for the large-scale production of propionic acid in the near future.
1. Introduction
Propionic acid is a 3-carbon short chain fatty acid with wide applications in food and dairy, pharmaceutical, cosmetic, and agro-industries. It is chiefly used as a mold inhibitor for animal feeds and as a preservative for foods such as cheeses and baked goods.1 In other fields, propionic acid can be used for animal therapy, and for the manufacture of perfume, herbicides, cellulose plastics, and flavoring agents.2 Currently, almost all propionic acid is synthesized commercially via petrochemical routes, with an annual world-wide market demand of more than 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 tons.3 However, as the crude oil prices had surpassed US$100 per barrel, and the demand by consumers and manufacturers for using bio-based natural ingredients in foods, cosmetics, and pharmaceuticals has grown, there has been increasing interest in propionic acid production from microbial fermentation. Propionibacteria have been granted “Generally Recognized As Safe (GRAS)” status by the United States Food and Drug Administration (FDA).4 Therefore, fermentation by Propionibacteria has good potential for the production of natural propionic acid to satisfy the market demand, and also opens the door to a more sustainable approach for propionic acid manufacturing in the case with limited petroleum supply.
000 tons.3 However, as the crude oil prices had surpassed US$100 per barrel, and the demand by consumers and manufacturers for using bio-based natural ingredients in foods, cosmetics, and pharmaceuticals has grown, there has been increasing interest in propionic acid production from microbial fermentation. Propionibacteria have been granted “Generally Recognized As Safe (GRAS)” status by the United States Food and Drug Administration (FDA).4 Therefore, fermentation by Propionibacteria has good potential for the production of natural propionic acid to satisfy the market demand, and also opens the door to a more sustainable approach for propionic acid manufacturing in the case with limited petroleum supply.
Several fermentation processes have been studied for bio-production of propionic acid by genus Propionibacteria from renewable and low-cost substrates, such as corn meal,5 cane molasses,6 Jerusalem artichoke hydrolysate,7 flour hydrolysate,8 and crude glycerol.9 In general, carbon source is usually used in relatively higher concentration compared to other medium components and thus has a high share in the raw material cost. Therefore, the continuous exploitation of cheap, renewable carbon sources has been strongly stimulated. Whey is a by-product of cheese and casein production and an inexpensive raw material which is composed mainly of 4.5–5% (w/v) lactose, 0.6–0.8% (w/v) soluble proteins, 0.4–0.5% (w/v) lipids and mineral salts (8–10% of dried extract).10 Much of the whey is currently used as animal feed, fertilizer and as a food ingredient or additive.11 Nevertheless, as much as 180 million tons of the annual world whey production remains underutilized.12 Because whey is considered an environmentally high-strength wastewater pollutant due to its high biological oxygen demand (∼50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg L−1) and chemical oxygen demand (∼80
000 mg L−1) and chemical oxygen demand (∼80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg L−1) contents, disposal of whey can be costly.13 Several other problems will also be encountered in whey disposal, including uneconomical transport due to its high water content and the difficulty of prolonged storage because of whey susceptibility to spoilage by microorganisms. Therefore, both environmental and economic concerns demand that whey should be converted into value-added products. To our knowledge, whey lactose has only been used in a few cases to produce propionic acid by Propionibacteria so far.14 However, the yield and productivity of propionic acid were not satisfactory for industrial application, resulting in difficulty in product recovery and decreased cost-effectiveness. It is thus of importance to develop an effective fermentation technology for economical production of propionic acid.
000 mg L−1) contents, disposal of whey can be costly.13 Several other problems will also be encountered in whey disposal, including uneconomical transport due to its high water content and the difficulty of prolonged storage because of whey susceptibility to spoilage by microorganisms. Therefore, both environmental and economic concerns demand that whey should be converted into value-added products. To our knowledge, whey lactose has only been used in a few cases to produce propionic acid by Propionibacteria so far.14 However, the yield and productivity of propionic acid were not satisfactory for industrial application, resulting in difficulty in product recovery and decreased cost-effectiveness. It is thus of importance to develop an effective fermentation technology for economical production of propionic acid.
Numerous attempts have been made to improve propionic acid fermentation, and have resulted in the development of innovative culture technologies and mutant strains.15 To date, the most promising bioprocess employs a fibrous-bed bioreactor (FBB) in a fed-batch cultivation mode with cells immobilized in a cotton towel packed in the reactor, which has been demonstrated to successfully produce several organic acids (acetic acid, propionic acid, lactic acid, and butyric acid) with significantly improved reactor productivity, final product yield and concentration.5,16–18 Despite the high final concentration obtained, the propionic acid fermentation was usually time-consuming (97.0 g L−1 in FBB at 2000 h and 67.1 g L−1 in MFB at 496 h of fermentation),17,19 which makes cotton fiber inefficient for the industrial production of propionic acid. In addition, cotton fiber has a small specific surface area and a low porosity, which not only reduces the number of immobilized cells on the fiber but also decreases its update rate. In the present work, production of propionic acid from whey lactose was investigated using cells of Propionibacterium acidipropionici immobilized on Poraver, one kind of porous recycled glass beads, treated with a cationic polymer, polyethylenimine.20 Immobilized-cell bioreactors were operated in batch and fed-batch fermentation modes, respectively, to determine process efficiency and economics.
Trehalose is a non-reducing disaccharide (1-O-a-dglucopyranosyl-α-D-glucopyranoside) naturally accumulated in microbes, insects, and plants. Trehalose has many advantages due to its inertness property and the ability to stabilize the biomolecules and production of this disaccharide is of commercial importance.21 Initially, trehalose was thought to act solely as a reserve energy and carbon source in a manner similar to that of starch and glycogen, but a growing number of studies indicate that this sugar instead has the important biological function of playing a major role in cell protection against various physical and chemical stresses, such as heat, cold, dehydration, desiccation, oxygen radicals, and acidic/alkali environmental conditions.22–24 Moreover, the ability of the microorganisms to survive in these environments correlates with their trehalose content.25,26 In recent years, trehalose was reported in cell extracts obtained from different species of the genus Propionibacteria, and the intracellular trehalose content increased considerably in response to osmotic, oxidative and acid stress.27–29 Inspired by this observation, the second goal was to elucidate the trehalose metabolism and regulation in P. acidipropionici, as well as its contribution to the enhancement of propionic acid resistance during the immobilization process in the FBB. We anticipated that the accumulation of trehalose engineered by strain manipulation could be a good strategy to improve the stress resistance and allow extended fermentation.
2. Materials and methods
2.1. Chemicals
Propionic acid, acetic acid, and succinic acid were all of analytical grade, and were purchased from Guoyao Reagent Co., Ltd (Shanghai, China). Whey powder was obtained from Anhui Huayang Farming Co., Ltd (Anhui province, China). Yeast extract and trypticase were purchased from Oxoid Ltd (Basingstoke, Hampshire, England). Poraver® beads (6–8 mm diameter) were obtained from Germany Marcom Co., Ltd (Beijing Office, China). Polyethylenimine (PEI, average Mw = 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and antibiotics were procured from Sigma-Aldrich (St. Louis, MO, USA). Other chemicals of analytical grade were obtained from standard sources.
000) and antibiotics were procured from Sigma-Aldrich (St. Louis, MO, USA). Other chemicals of analytical grade were obtained from standard sources.
2.2. Preparation of whey lactose
Two hundred grams of whey powder (spray dried powder containing lactose >65% (w/v) and protein >11% (w/v)) was added to 100 mL of sodium phosphate buffer (pH 7.5) and mixed vigorously. The mixture was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min, and the supernatant was adjusted to pH 7.5. The mixture was centrifuged again at 12
000g for 20 min, and the supernatant was adjusted to pH 7.5. The mixture was centrifuged again at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min. The lactose concentration of the supernatant was approximately 20% (w/v) and the prepared lactose solution was stored at 4 °C for further use.30
000g for 20 min. The lactose concentration of the supernatant was approximately 20% (w/v) and the prepared lactose solution was stored at 4 °C for further use.30
2.3. Strains and media
Propionibacterium acidipropionici CGMCC 1.2232 (equal to Propionibacterium acidipropionici ATCC 4875) was purchased from the China General Microbiological Culture Collection Center (CGMCC) and designated as the wild-type. Escherichia coli strains DH5α and JM109 (λ pir) were used for the cloning, maintenance and propagation of plasmids. E. coli S17–1 (λ pir) was used to transfer plasmid DNA via conjugation to P. acidipropionici. The primers used in this work are displayed in Table S1 in the ESI.†E. coli strains were grown at 37 °C aerobically in Luria-Bertani (LB) medium. When necessary, antibiotics were added at the following final concentrations: 50 μg mL−1 each for ampicillin, kanamycin, streptomycin and erythromycin, and 25 μg mL−1 for spectinomycin. The stock culture of P. acidipropionici was kept in serum bottles under anaerobic conditions at 4 °C.31 For P. acidipropionici, a detailed description of the culture medium has been given elsewhere.7 Acid stress was imposed by the addition of 100 g L−1 propionic acid to the culture medium. The basal culture medium (without the carbon source) and the concentrated carbon source solution were sterilized separately by autoclaving at 121 °C and 15 psig for 20 min to avoid undesirable reactions.32 They were mixed aseptically before use in the fermentation study.
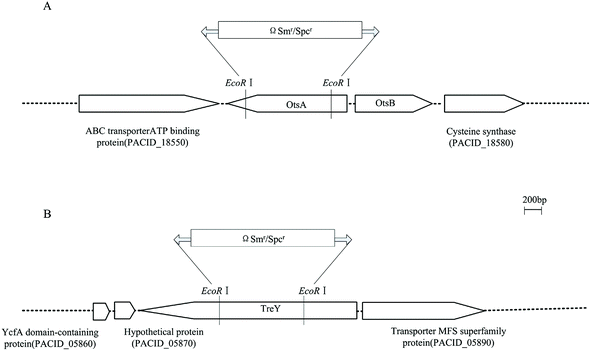
2.4. Construction of ΔotsA, ΔtreY and ΔotsAΔtreY mutants of P. acidipropionici
Plasmid isolation, restriction enzyme digestion, DNA ligation, and the transformation of E. coli were performed as described by Sambrook and Russell.33 The PCR primers used in this work are displayed in Table 1. A 1.0 kb DNA fragment with the 5′ region of the otsA gene was generated using the otsA_f1 primer and otsA_r1 primer with P. acidipropionici DNA by the PCR reaction. This DNA fragment was digested and ligated with XbaI–EcoRI digested pBluescriptII SK+ (Stratagene), resulting in pBOTSAI. A 1.1 kb DNA fragment with the 3′ region of the otsA gene was generated using the otsA_f2 primer and otsA_r2 primer. This DNA fragment was digested, ligated with EcoRI–HindIII digested pBOTSAI, resulting in pBOTSAII. The 2.0 kb Smr/Spcr Ω cassette was inserted into the EcoRI site of pBOTSAII to create pBOTSAW. It is flanked by short inverted repeats with the T4 transcription-termination signal. The 4.1 kb XbaI–KpnI restriction fragment from pBOTSAW was inserted into the Kmr-suicide vector pJP5603, which predigested with XbaI and KpnI, resulting in pJPOTSA. An 1.0 kb DNA fragment with the 5′ region of the treY gene was generated using the treY_f1 primer and treY_r1 primer by PCR. Then it was digested, ligated with XbaI–EcoRI digested pBluescriptII SK+, resulting in pBTREYI. A 1.2 kb DNA fragment with the 3′ region of the treY gene was generated using the treY_f2 primer and treY_r2 primer. This fragment was digested and then ligated with digested pBTREYI by EcoRI–HindIII, resulting in pBTREYII. The Smr/Spcr Ω cassette was inserted into the EcoRI site of the pBTREYII to create pBTREYW. The 4.2 kb XbaI–KpnI restriction fragment from pBTREYW was inserted into the XbaI and KpnI sites of pJP5603 to create pJPTREY. Then the plasmid was transferred into P. acidipropionici cells by conjugation with mobilizing strain E. coli S17-1 (λ pir). Spcr Kms cells were selected as double-crossover candidates, and the chromosomal insertion was confirmed by PCR amplification and DNA sequencing. To generate strains carrying double gene deletion, single deletion strain was used instead of the wild-type. A two-step recombination system was used for inactivating the trehalose biosynthesis genes of P. acidipropionici to make the double mutant strain ΔotsAΔtreY described elsewhere.34| Designation | Oligonucleotide sequence (5′–3′) | Digestion enzyme |
|---|---|---|
| otsA_f1 | ATA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ATG TCC GAA CCC GTT CAG G ATG TCC GAA CCC GTT CAG G |
XbaI |
| otsA_r1 | TAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GGC CGA CCA GCA GGT CGA T GGC CGA CCA GCA GGT CGA T |
EcoRI |
| otsA_f2 | ATA![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ACG ACT GCA TCG CCG AAC C ACG ACT GCA TCG CCG AAC C |
EcoRI |
| otsA_r2 | TAT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) TCA CTC GCC CTC CAG CTG G TCA CTC GCC CTC CAG CTG G |
HindIII |
| treY_f1 | ATA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GTG GTC GTG ACG TCC TCC AC GTG GTC GTG ACG TCC TCC AC |
XbaI |
| treY_r1 | TAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) CGA GGC GCT CCC AGA TGT C CGA GGC GCT CCC AGA TGT C |
EcoRI |
| treY_f2 | ATA![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) CCG GCC CGA CGG TGC C CCG GCC CGA CGG TGC C |
EcoRI |
| treY_r2 | TAT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) TCA GTC CGC CAC GAG AAG G TCA GTC CGC CAC GAG AAG G |
HindIII |
2.5. Construction of over-expression mutant of P. acidipropionici
To overexpress otsA gene in the OtsA–OtsB pathway, the 1.82 kb full-length otsA gene from P. acidipropionici CGMCC 1.2232 was amplified by PCR using genomic DNA as the template and the oligonucleotide primers otsA_f (5′-ATA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ATGTCCGAACCCGTTCAGG-3′) and otsA_r (5′-TAT
ATGTCCGAACCCGTTCAGG-3′) and otsA_r (5′-TAT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TCACTCGCCCTCCAGCTGGT-3′). The PCR product was ligated into the pGEM-T Easy vector to create pGEM-otsAEX. The pGEM-otsAEX was subsequently digested with BclI and SacII, and the 1.82 kb fragment containing otsA was inserted into the BclI and SstII sites of the shuttle vector pBRESP36A, yielding plasmid pBRESP36A-otsA. Expression of genes inserted into this vector was constitutively with the Eryr and Ampr genes.35
TCACTCGCCCTCCAGCTGGT-3′). The PCR product was ligated into the pGEM-T Easy vector to create pGEM-otsAEX. The pGEM-otsAEX was subsequently digested with BclI and SacII, and the 1.82 kb fragment containing otsA was inserted into the BclI and SstII sites of the shuttle vector pBRESP36A, yielding plasmid pBRESP36A-otsA. Expression of genes inserted into this vector was constitutively with the Eryr and Ampr genes.35
2.6. Fermentation kinetic studies with fibrous bed bioreactor
The fermentation system consisted of a 5 L stirred-tank fermentor (B. Braun, B. Braun Biotech International, Melsungen, Germany) connected to a 0.5 L FBB through a recirculation loop and operated at 32 °C, agitated at 150 rpm, and the pH was controlled at 6.0 by adding NH4·OH facilitated by an on-line sensing and dosing system.36 The immobilization matrices of Poraver coated with PEI were prepared as described elsewhere.20 The batch and fed-batch modes were started with 2 L of fresh medium, and the fed-batch mode was followed with pulse feeding concentrated whey lactose whenever its concentration in the fermentation broth was close to zero. The results shown are the mean of analyses performed in duplicates for all the fermentations.2.7. Trehalose determination
To determine the intracellular trehalose level in P. acidipropionici, 10 mL of broth from the culture medium was collected at regular intervals of time and centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 4 °C. A few grams of the pellets obtained were then extracted with 80% ethanol (5 ml per 1 g wet weight), suspended and homogenized well. The suspension was boiled at 100 °C for 5 min and centrifuged at 16
000g for 10 min at 4 °C. A few grams of the pellets obtained were then extracted with 80% ethanol (5 ml per 1 g wet weight), suspended and homogenized well. The suspension was boiled at 100 °C for 5 min and centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min. The supernatant was filtered with a Minisart-RC 4 filter (Sartorius) and added with phosphate buffer (100 mM, pH 5.7) to make the final volume to 1 mL. Trehalose was established by assaying the release of free glucose following incubation of samples with trehalase (EC 3.2.1.28; Sigma-Aldrich).37 Samples for analysis (100 μL) were added to 100 μL phosphate buffer (100 mM, pH 5.7), 800 μL water and 10 μL trehalase, then incubated at 37 °C for 1 h and stopped by incubation in boiling water for 5 min. Glucose was then measured using a HPLC system as described below.
000g for 10 min. The supernatant was filtered with a Minisart-RC 4 filter (Sartorius) and added with phosphate buffer (100 mM, pH 5.7) to make the final volume to 1 mL. Trehalose was established by assaying the release of free glucose following incubation of samples with trehalase (EC 3.2.1.28; Sigma-Aldrich).37 Samples for analysis (100 μL) were added to 100 μL phosphate buffer (100 mM, pH 5.7), 800 μL water and 10 μL trehalase, then incubated at 37 °C for 1 h and stopped by incubation in boiling water for 5 min. Glucose was then measured using a HPLC system as described below.
2.8. Analysis methods
Cell growth was analyzed by measuring the optical density of the cell suspension at a wavelength of 600 nm (OD600) with a spectrophotometer (Ultrospec 3300 pro, Amersham Bioscience) after an appropriate dilution. Dry cell weight (DCW) was calculated from the OD value using a correlated equation (DCW (g L−1) = 0.64 × OD600).36 Quantitative analysis of lactose, glucose and organic acid products (propionic, acetic, and succinic acids) in the fermentation broth was performed using a HPLC system equipped with an organic acid column (Bio-Rad, HPX-87H). The HPLC analysis was operated at 45 °C using 5 mM H2SO4 as the eluant, with a flow rate of 0.6 mL min−1. All results were obtained from the means of triplicate determinations.3. Results and discussion
P. acidipropionici was chosen as a candidate for propionic acid fermentation based on earlier reports which showed that the bacterium provided a higher conversion yield and productivity among the Propionibacteria spp. investigated,7,19,38 including fermentation on a semi-industrial scale (10 m3),39 as well as an available genome sequence.31 Lactose from cheese whey is superior to other carbon sources because of its low market price with a comparable level of propionic acid yield. Theoretically, its metabolism yields 0.79 gram propionic acid per gram of lactose; however, under experimental conditions, the yield ranges from 0.40 to 0.70 g per g lactose with succinic acid and acetic acid as the major by-product.403.1. Batch fermentation of whey lactose with free cells
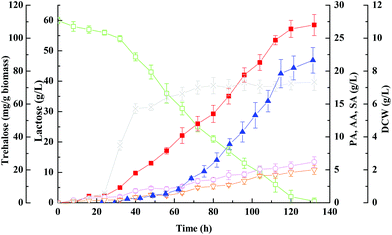
The kinetics of propionic acid batch fermentation of whey lactose using free P. acidipropionici cells was illustrated in Fig. 1. Lactose (60 g L−1) was entirely consumed in 132 ± 10 h yielding 27 ± 1.6 g per L propionic acid at a productivity of ∼0.20 g (L h)−1. Total organic acids yield from lactose was 0.63 g g−1, 70.7% (g g−1) of which was propionic acid. As shown in Fig. 1, the free-cell fermentation was characterized by a very long lag phase of about 25 h, and production of organic acids was minimal in this period but increased afterward and reached peak values. However, when the propionic acid concentration reached up to 20 g L−1, the growth rate approximated to zero, and cells were still metabolically active and continued to produce propionic acid at a lower rate.7 As a weak organic acid, the non-dissociated propionic acid can pass through the cell membrane into the cytoplasm and release protons due to the intracellular alkaline environment that results in disturbing of the pH gradient across the cell membrane and affects the nutrient transfer and inhibits cell growth.173.2. Batch fermentation with cell immobilized on PEI-Poraver
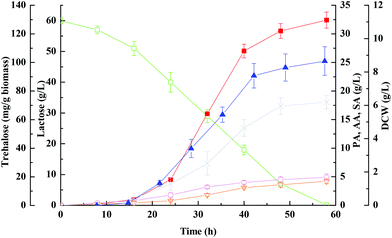
Poraver is a trade name for foamed highly porous recycled glass beads, which has been studied previously in biofilm reactors.41 Its surface is covered by numerous macropores which increase the surface area and support cell attachment and settlement. However, native Poraver beads did not exhibit significant adsorption of microorganisms even after a few days incubation with the cells (data not shown). Based on adsorption studies, cell adsorption using the charged cationic polymer polyethylenimine (PEI) is an extremely simple and straightforward procedure resulting in the negatively charged cells becoming firmly attached to the matrix. Cell immobilization was performed using the procedure described earlier by us for adsorbing Clostridium tyrobutyricum on PEI coated cotton fiber,42 with the exception that the cells were grown to high density in the reactor vessel before circulation through the column packed with the matrix. This helped to overcome the inhibitory effect of PEI exerted on cell growth and to shorten the immobilization period.As a result, the batch fermentation of whey lactose with immobilized cells on PEI-Poraver in FBB was illustrated in Fig. 2. Compared with those under free-cell fermentation conditions (Fig. 1), bacteria immobilized on PEI-Poraver grew faster and exhibited more lactose consumption, a shorter fermentation period, and a faster rate of propionic acid formation in the FBB. With an initial lactose concentration of 60 g L−1, the maximum propionic acid concentration reached 32.4 ± 1.4 g L−1 at 58 ± 4 h in the FBB (productivity ∼ 0.56 g (L h)−1), which showed that whey lactose could be efficiently metabolized by immobilized P. acidipropionici, in accordance with the previous reports.43,44 A comparable observation was also recorded by Coral et al.45 in processes with both glycerol and lactose as culture media carbon sources. At ∼130 h of batch fermentation, lactose introduced with medium was exhausted while half of glycerol initial amount remained. Moreover, the amount of organic acid by-products of acetic acid and succinic acid were also reduced in the FBB, which could be beneficial to the separation and purification of propioinc acid in industrial production.
3.3. Fed-batch fermentation with immobilized cells
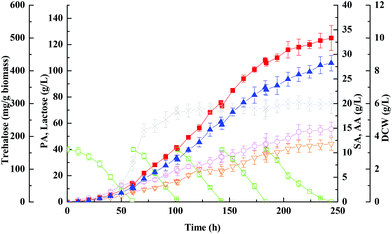
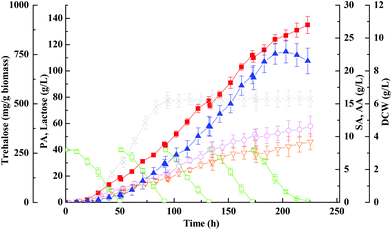
The use of concentrated lactose solutions (up to 200 g L−1) is important in an actual industrial production process since it will allow high propionic acid titres (up to 10–15% v/v) at the end of fermentation, therefore further reducing considerably the propionic acid distillation costs. However, natural lactose-fermenting Propionibacteria cannot ferment efficiently (i.e., rapidly and with high conversion yields) such high concentrations of lactose. Since fed-batch fermentation can alleviate the inhibition of high sugar concentration and minimize the loss of the remaining substrate, but maximize the final concentration and yield of a target product, it is often favored over batch fermentation. Based on the previous findings, the fed-batch fermentation approach with immobilized cells on PEI-Poraver was employed here to improve the final concentration of propionic acid from whey lactose (Fig. 3). As can be seen in Fig. 3, Propionibacteria grew exponentially in the first fed-batch and then entered the stationary phase in the middle of the second batch. It continued to produce propionic acid until the final concentration reached 125 ± 9.6 g L−1 within the whole period of ∼250 h, which was much higher than that obtained in the batch fermentation (32.4 ± 1.4 g L−1). Another further positive effect of the fed-batch supply was the higher selectivity of propionic acid production discussed above, which is, the ratio of propionic acid to total organic acids (g g−1) was increased from batch culture to fed-batch culture (82.5% vs. 78.1%), a clear indication that the metabolic pathway in the bacterial had been shifted. Chen et al. proved through metabolic flux analysis that the fluxes of propionic acid synthesis and the pentose phosphate pathway in immobilized-cell fermentation increased by 84.7% and 228% compared with free cell fermentation, and also proved a decrease in succinic and acetic acid fluxes.46 However, the productivity decreased dramatically at the propionic acid concentration higher than 100 g L−1, and at the end of fermentation productivity reached 0.50 g (L h)−1, which decreased by 10.7% compared with the batch fermentation, indicating a strong product inhibition at this level (Table 2).| Culture | Free-cell batch fermentation with wild-type strain | FBB with immobilized wild-type strain | Free-cell batch fermentation with otsA over-expression strain | Fed-batch FBB with immobilized otsA over-expression strain | |
|---|---|---|---|---|---|
| Batch fermentation | Fed-batch fermentation | ||||
| a Mean ± standard deviation (n = 3). b Biomass of free cells in 5 L fermenter. c Based on the assumption that 46.2% of the biomass was carbon. The production of CO2 was calculated based on the assumption that one mole of CO2 was co-produced with each mole of acetic acid in the fermentation according to the metabolic pathways in P. acidipropionici.16 | |||||
| Lactose (g L−1) | 60 ± 1.2 | 60 ± 1.8 | 200 ± 8.5 | 60 ± 1.4 | 200 ± 7.2 |
| Product concentration (g L−1) | |||||
| Propionic acid | 27 ± 1.6 | 32.4 ± 1.4 | 125 ± 9.6 | 30 ± 1.7 | 135 ± 6.5 |
| Acetic acid | 6.2 ± 0.8 | 4.9 ± 0.6 | 14.8 ± 1.4 | 5.6 ± 0.3 | 11.6 ± 1.6 |
| Succinic acid | 5.0 ± 0.7 | 4.2 ± 0.4 | 11.8 ± 1.3 | 4.6 ± 0.4 | 9.2 ± 1.7 |
| Biomass concentration (g L−1) | 7.3 ± 0.5 | 6.2 ± 0.4b | 6.0 ± 0.6b | 7.1 ± 0.6 | 6.3 ± 0.5b |
| Product yield (g g−1) | |||||
| Propionic acid | 0.45 ± 0.028 | 0.54 ± 0.022 | 0.62 ± 0.048 | 0.50 ± 0.041 | 0.67 ± 0.067 |
| Acetic acid | 0.10 ± 0.01 | 0.082 ± 0.018 | 0.074 ± 0.007 | 0.093 ± 0.008 | 0.058 ± 0.010 |
| Succinic acid | 0.083 ± 0.008 | 0.070 ± 0.013 | 0.059 ± 0.007 | 0.076 ± 0.009 | 0.046 ± 0.010 |
| Biomass yield (g g−1) | 0.12 ± 0.017 | 0.10 ± 0.017 | 0.03 ± 0.007 | 0.12 ± 0.011 | 0.032 ± 0.003 |
| Productivity (g (L h)−1) | 0.20 ± 0.034 | 0.56 ± 0.075 | 0.50 ± 0.10 | 0.25 ± 0.038 | 0.61 ± 0.10 |
| Propionic acid/total acids (%, g g−1) | 70.7 ± 4.5 | 78.1 ± 4.8 | 82.5 ± 5.2 | 74.6 ± 4.8 | 86.6 ± 7.4 |
| Total carbon recovery (%)c | 87.9 ± 7.5 | 92.0 ± 4.7 | 91.7 ± 5.6 | 91.4 ± 6.6 | 94.0 ± 4.2 |
| Fermentation time (h) | 132 ± 10 | 58 ± 4 | 250 ± 12 | 120 ± 10 | 220 ± 12 |
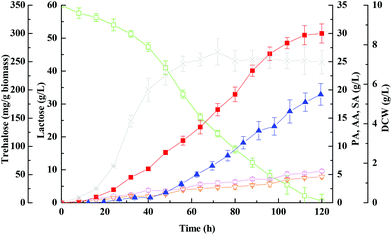
The common defence strategy of microbes against environmental stresses includes accumulation of several organic compatible solutes.47 Realizing the importance of trehalose as a well-known compatible solute accumulated in microbes, it is not surprising to find that the cells accumulated abundant trehalose in response to increased propionic acid concentration in the culture medium (Fig. 1 and 2), especially up to 100 g L−1 (Fig. 3). As can be seen in Fig. 3, the maximum trehalose yield with respect to biomass was 423 ± 23 mg g−1, which was more than 3 times higher than that obtained with pure glycerol (128 ± 3 mg g−1) by genus Propionibacteria.9 Thus it can be predicted that the presence of certain other impurities in whey except for lactose is probably responsible for the enhancement of trehalose production.
Previously, enhanced trehalose yield under environmental stress (e.g., osmotic stress) was reported in microbes, including P. freudenreichii, E. coli, C. glutamicum and yeast.27,48–50 Further experiments were thus conducted to search the genome sequence of P. acidipropionici with homology to genes involved in trehalose biosynthesis in other bacteria, and constructed mutants using gene inactivation and over-expression of various enzymes in the trehalose biosynthesis metabolic pathway to compare their phenotypes and regulatory properties under acid stress with those of the wild-type strain.
3.4. Trehalose synthesis pathway in P. acidipropionici
The availability of complete genome sequence data of P. acidipropionici CGMCC 1.2232 (http://www.ncbi.nlm.nih.gov/genome/13350?project_id=179069) makes it possible to screen for the presence of ORFs sharing homology with genes known to be involved in trehalose biosynthesis in other bacteria. The ORFs with high similarity to four genes involved in two different pathways were found in P. acidipropionici and the results are summarized in Table 3. A BLAST search based on the known E. coli OtsA–OtsB pathway genes identified ORFs PACID18560 and PACID18570 as a homologue of trehalose 6-phosphate synthase (OtsA) and trehalose 6-phosphate phosphatase (OtsB) as having 36% and 31% overall identity at the amino acid level respectively. The ORFs PACID05880 and PACID05850 were identified as treY and treZ, respectively, because they have 39% and 46% identity with the Rhizobium sp. TreY–TreZ pathway genes.| P. acidipropionici ORFs | Length (aa) | Matching sequences | Length (aa) | Identity (%) | |
|---|---|---|---|---|---|
| Strain | Description | ||||
| OtsA (putative) (PACID:18560) | 467 | Escherichia coil | Trehalose 6-phosphate synthase OtsA | 474 | 36% |
| Salmonella typhimurium | Trehalose 6-phosphate synthase OtsA | 473 | 37% | ||
| Saccharomyces cerevisiae | Trehalose 6-phosphate synthase TPS1 | 495 | 36% | ||
| OtsB (putative) (PACID:18570) | 301 | Escherichia coil | Trehalose 6-phosphate synthase OtsB | 266 | 31% |
| Salmonella typhimurium | Trehalose 6-phosphate synthase OtsB | 267 | 31% | ||
| Saccharomyces cerevisiae | Trehalose 6-phosphate synthase TPS2 | 896 | 23% | ||
| TreY (putative) (PACID:05880) | 847 | Rhizobium sp. | Maltooligosyl trehalose synthase TreY | 772 | 39% |
| Arthrobacter sp. | Maltooligosyl trehalose synthase TreY | 775 | 38% | ||
| TreZ (putative) (PACID:05850) | 584 | Rhizobium sp. | Maltooligosyl trehalose synthase TreZ | 596 | 46% |
| Arthrobacter sp. | Maltooligosyl trehalose synthase TreZ | 598 | 46% | ||
In order to characterize the role of two distinct trehalose-biosynthesis pathways proposed from genome sequence data analysis in P. acidipropionici, two mutants were constructed by chromosomal inactivation of one gene of each pathway, and named ΔotsA and ΔtreY respectively (Fig. 4). Based on the single mutants, the combination of a double mutant named ΔotsAΔtreY was constructed by the two-step recombination system. Also, over-expression was carried out via a replicative plasmid pGEM-T containing otsA gene in the OtsA–OtsB pathway from P. acidipropionici. First, we measured the intracellular trehalose level among P. acidipropionici wild-type and trehalose biosynthesis gene mutants under acid stress with culture media containing 100 g L−1 propionic acid. As can be seen in Table 4, the ΔotsA mutant showed a significant decrease in the intracellular trehalose level in comparison with the wild-type strain (95 ± 10 vs. 311 ± 17 mg g−1 biomass), while the level in the ΔtreY mutant also decreased to some extent (230 ± 12 vs. 311 ± 17 mg g−1 biomass). The results led to the proposal that trehalose synthesis in P. acidipropionici cells responding to acid stress predominantly depends on the OtsA–OtsB pathway and only marginally on the TreY–TreZ pathway, which were in accordance with those for R. sphaeroides, but inconsistent with those for C. glutamicum.34,48 For example, in the later organism, it has been shown that the mutant with a deletion of the otsA gene accumulates levels of trehalose comparable with those of the wild-type, with the synthesis proceeding via the TreY–TreZ pathway, which revealed an enormous disparity in the function of trehalose synthesis pathways. However, there was no detectable intracellular trehalose in the ΔotsAΔtreY mutant strain, and experiments with otsA over-expression mutant strain found a significant increase in the intracellular trehalose level (up to 566 ± 29 mg g−1 biomass), which was 1.8 times higher than the value in wild-type strain.
| Strains | Trehalose yield (mg per g biomass) |
|---|---|
| a The trehalose levels were determined at 80 h. All data shown are mean values from at least three replicate experiments. | |
| P. acidipropionici wild type | 311 ± 17 |
| ΔotsA | 95 ± 10 |
| ΔtreY | 230 ± 12 |
| ΔotsAΔtreY | 0 |
| otsA over-expression mutant | 566 ± 29 |
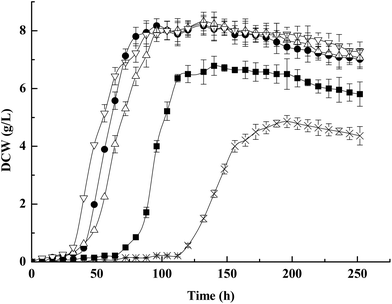
We next examined the growth characteristics under acid-stressed conditions (100 g per L propionic acid), and the significant difference in growth curve of strains was observed (Fig. 5). As can be seen in Fig. 5, the growth was slightly compromised in both ΔotsA and otsA over-expression mutant strains and strongly compromised in the ΔotsAΔtreY mutant strain compared to the wild-type strain. A wild-type strain reached the maximum DCW about 8.06 g L−1, while ΔotsA and otsA over-expression mutant strains reached the maximum DCW about 6.78 g L−1 and 8.11 g L−1, respectively. Additionally, the ΔotsAΔtreY mutant strain reached the maximum DCW about 4.85 g L−1. The results showed that the acid-stressed conditions showed a significant growth suppression in the ΔotsAΔtreY mutant strain compared to the wild-type strain. However, there was a recovery of acid tolerance in the ΔotsAΔtreY mutant strain when adding 10 mM trehalose to the acid-containing-medium (Fig. 5), and no remarkable growth difference was observed between the wild-type and ΔotsAΔtreY mutant strain under this condition, which indicates that the growth suppression was caused by impossibility of trehalose synthesis. Previous research has revealed that accumulation of constitutive trehalose in E. coli under stress conditions is thought to contribute to maintenance of cell integrity and protein stability.50 Consistent with such a role, the presence of free trehalose in apparently acid-stressed Propionibacteria may likewise have the same protective function.
3.5. Production of propionic acid with enhanced trehalose synthesis
Concerning the growth profiles between wild-type and mutants, as well as the finding that the OtsA–OtsB pathway is the major route for trehalose synthesis under acid stress, the otsA over-expression mutant strain with enhanced trehalose synthesis was further used to improve propionic acid production. Fig. 6 and 7 show the free-cell batch and immobilized-cell fed-batch fermentation kinetics with the otsA over-expression mutant, respectively. The results were further compared with the wild-type strain listed in Table 2. As can be seen in Fig. 6, by-products acetic acid and succinic acid yield in otsA over-expression mutant declined very slightly in constrast to the wild-type in the free-cell fermentation process. In contrast, the propionic acid yield with the mutant was 11% higher (0.50 ± 0.041 vs. 0.45 ± 0.028) than that with the wild-type strain. In the immobilized-cell fermentation mode shown in Fig. 7, the otsA over-expression mutant strain produced 21% less acetic acid (0.058 ± 0.010 vs. 0.074 ± 0.007 g g−1) and 22% less succinic acid (0.046 ± 0.010 vs. 0.059 ± 0.007 g g−1) than the wild-type strain. In contrast, the propionic acid yield with the otsA over-expression mutant strain was 11% higher (0.50 ± 0.041 vs. 0.45 ± 0.028) than that with the wild-type strain. However, the over-expression mutant had a higher propionic acid production performance in FBB fed-batch fermentation mode even in the first batch. The maximum concentration of propionic acid reached 135 ± 6.5 g L−1 with a corresponding productivity and yield of 0.61 ± 0.10 g (L h)−1 and 0.67 ± 0.067 g g−1 lactose, respectively, which to our knowledge is among the highest propionic acid concentration ever produced in the traditional fermentation process. So far, the maximum propionic acid concentration previously produced in fermentation was 136.23 ± 6.77 g L−1 by P. freudenreichii CCTCC M207015 with glucose as the substrate.46 Zhang and Yang (2009) also reported that 97 g L−1 of propionic acid was produced from glucose by immobilized cells of P. acidipropionici in fed-batch fermentation.17 The higher propionic acid concentration obtained in these fed-batch immobilized fermentations indicated that the bacteria in the FBB had adapted and acquired a higher tolerance to propionic acid.Herein we showed that the enhancement of the OtsA–OtsB pathway in P. acidipropionici was a response common to the high acid concentration stress imposed. As expected, P. acidipropionici accumulated intracellular trehalose to remarkable levels, 20 hours in advance as compared to the wild-type strain. As shown in Fig. 7, its content increased considerably in response to acid accumulation, and the highest yield based on biomass consumed was improved 1.8 times over that in the wild-type strain (764 ± 47 mg g−1vs. 423 ± 14 mg g−1). Recent studies have demonstrated the ability of trehalose in cell protection against diverse stresses, such as osmotic stress, oxidative stress, heat stress as well as acid stress.51 It is conceivable that trehalose plays a role in the stabilization of cell membranes, which must withstand a strong pH gradient in such acidic environments. In addition, it also can be seen in Fig. 7 that there was a slow decrease in trehalose concentration with growth in the late stationary phase, which may be ascribed to the degradation of trehalose by hydrolytic enzymes. However, trehalase, trehalose phosphorylase, trehalose-6-phosphate phosphorylase and trehalose-6-phosphate hydrolase identified in other organisms involved in trehalose degradation are all absent in the genome sequence of P. acidipropionici, indicating a new catabolism pathway of trehalose in this food microbe.
4. Conclusions
Herein, we demonstrated the advantages of using immobilized cells for propionic acid fermentation from whey lactose to achieve the desired improvement in fermentation efficiency. In particular, immobilization on PEI-Poraver considerably enhanced propionic acid properties with both final propionic acid productivity (0.56 ± 0.075 g (L h)−1vs. 0.29 ± 0.034 g (L h)−1 with free-cell fermentation) and yield (0.54 ± 0.022 g g−1vs. 0.45 ± 0.028 g g−1 with free-cell fermentation). In addition, P. acidipropionici accumulated intracellular trehalose to remarkable levels, and its content increased considerably in response to acid accumulation. Two pathways (OtsA–OtsB, TreY–TreZ) to synthesize trehalose were identified in P. acidipropionici, and their functions were then studied by characterizing strains defective in individual trehalose biosynthetic routes. A phenotypic comparison revealed that trehalose synthesis in P. acidipropionici is mediated mainly by the OtsA–OtsB pathway and, to some extent, by the TreY–TreZ pathway. Strains with the simultaneous inactivation of otsA and treY genes belonging to these two pathways were completely unable to synthesize trehalose. On the other hand, the mutant strain with over-expression of the otsA gene could significantly increase the synthesis of trehalose up to 764 ± 47 mg g−1 biomass, which then promoted the production of propionic acid. The highest propionic acid concentration of 135 ± 6.5 g L−1 was superior to those reported earlier in either batch or fed-batch modes of operation with equivalent productivities. To further improve the final concentration of propionic acid, a deeper and global profile of the trehalose assimilation and metabolism in P. acidipropionici should be performed in further trials.Acknowledgements
This work was obtained from the National Science Foundation for Distinguished Young Scholars of China (no. 21225626), the National Basic Research Program of China (no. 2012CB725204), the National Natural Science Foundation of China for Young Scholars (no. 21106064), the Natural Science Foundation of Jiangsu Province (no. BK20131406, BK20130917), and the National High Technology Research and Development Program of China (no. 2012AA022101, 2012AA021700).References
- E. Maier, K. Kurz, M. Jenny, H. Schennach, F. Ueberall and D. Fuchs, Food Chem. Toxicol., 2010, 48, 1950–1956 CrossRef CAS PubMed.
- S. H. Al-Lahham, M. P. Peppelenbosch, H. Roelofsen, R. J. Vonk and K. Venema, Biochim. Biophys. Acta, 2010, 1801, 1175–1183 CrossRef CAS PubMed.
- C. C. Stowers, B. M. Cox and B. A. Rodriguez, J. Ind. Microbiol. Biol., 2014, 5, 837–852 CrossRef PubMed.
- D. A. Brede, T. Faye, M. P. Stierli, G. Dasen, A. Theiler, I. F. Nes, L. Meile and H. Holo, Appl. Environ. Microbiol., 2005, 71, 8077–8084 CrossRef CAS PubMed.
- Y. L. Huang, Z. T. Wu, L. K. Zhang, C. Ming Cheung and S. T. Yang, Bioresour. Technol., 2002, 82, 51–59 CrossRef CAS.
- X. H. Feng, F. Chen, H. Xua, B. Wu, H. Lia, S. Lia and P. K. Ouyang, Bioresour. Technol., 2011, 102, 6141–6146 CrossRef CAS PubMed.
- Z. X. Liang, L. Li, S. Li, Y. H. Cai, S. T. Yang and J. F. Wang, Bioprocess Biosyst. Eng., 2012, 35, 915–921 CrossRef CAS PubMed.
- W. Sabra, D. Dietz and A. P. Zeng, Appl. Microbiol. Biotechnol., 2013, 97, 5771–5777 CrossRef CAS PubMed.
- R. Ruhal, S. Aggarwal and B. Choudhury, Green Chem., 2011, 13, 3492–3498 RSC.
- P. M. R. Guimarães, J. A. Teixeira and L. Domingues, Biotechnol. Adv., 2010, 28, 375–384 CrossRef PubMed.
- A. Kassaa, M. Brownbridgeb, B. L. Parkera, M. Skinnera, V. Goulia, S. Goulia, M. Guoc, F. Lee and T. Hataa, Mycol. Res., 2008, 112, 583–591 CrossRef PubMed.
- S. Alonso, M. Rendueles and M. Díaz, Bioresour. Technol., 2011, 102, 9730–9736 CrossRef CAS PubMed.
- M. M. Abboud, I. H. Aljundi, K. M. Khleifat and S. Dmour, Biochem. Eng. J., 2010, 48, 166–172 CrossRef CAS PubMed.
- J. Morales, J. S. Choi and D. S. Kim, Environ. Prog., 2006, 25, 228–234 CrossRef CAS.
- V. Goswami and A. K. Srivastava, Appl. Microbiol. Biotechnol., 2001, 56, 676–680 CrossRef CAS.
- S. Suwannakham and S. T. Yang, Biotechnol. Bioeng., 2005, 91, 325–337 CrossRef CAS PubMed.
- A. Zhang and S. T. Yang, Biotechnol. Bioeng., 2009, 104, 766–773 CAS.
- L. Jiang, S. Li, Y. Hu, Q. Xu and H. Huang, Biotechnol. Bioeng., 2012, 109, 708–718 CrossRef CAS PubMed.
- X. H. Feng, F. Chen, H. Xu, B. Wu, J. Yao, H. J. Ying and P. K. Ouyang, Bioprocess Biosyst. Eng., 2010, 33, 1077–1085 CrossRef CAS PubMed.
- T. Dishisha, M. T. Alvarez and R. H. Kaul, Bioresour. Technol., 2012, 118, 553–562 CrossRef CAS PubMed.
- A. D. Elbein, Y. T. Pan, I. Pastuszak and D. Carroll, Glycobiology, 2003, 13, 17–27 CrossRef PubMed.
- O. Kandror, A. DeLeon and A. L. Goldberg, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9727–9732 CrossRef CAS PubMed.
- J. E. Purvis, L. P. Yomano and L. O. Ingram, Appl. Environ. Microbiol., 2005, 71, 3761–3769 Search PubMed.
- L. Jiang, M. Lin, Y. Zhang, Y. Li, X. Xu and H. Huang, PLoS One, 2013, 8, e77437 CAS.
- A. D. Q. Nguyen, Y. G. Kim, S. B. Kim and C. J. Kim, Bioresour. Technol., 2013, 143, 531–537 CrossRef CAS PubMed.
- J. H. Crowe, F. A. Hoekstra and L. M. Crow, Annu. Rev. Physiol., 1992, 54, 579–599 CrossRef CAS PubMed.
- F. S. Cardosoa, P. Gaspara, J. Hugenholtzb, A. Ramosa and H. Santos, Int. J. Food Microbiol., 2004, 91, 195–204 CrossRef.
- F. S. Cardoso, R. F. Castro, N. Borges and H. Santos, Microbiology., 2007, 153, 270–280 CrossRef CAS PubMed.
- R. Ruhal and B. Choudhury, Bioresour. Technol., 2012, 109, 131–139 CrossRef CAS PubMed.
- Y. S. Songa, H. Y. Shina, J. Y. Lee, C. Park and S. W. Kim, Food Chem., 2012, 133, 611–617 CrossRef PubMed.
- L. Huang, Y. J. Xiang, J. Cai, L. Jiang, Z. B. Lv, Y. Z. Zhang and Z. N. Xu, Korean J. Chem. Eng., 2011, 28, 2312–2315 CrossRef CAS.
- L. Jiang, J. F. Wang, S. Z. Liang, J. Cai, Z. N. Xu, P. L. Cen, S. T. Yang and S. Li, Biotechnol. Bioeng., 2011, 108, 31–40 CrossRef CAS PubMed.
- J. Sambrook and D. W. Russell, Molecular cloning: a laboratory manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 3rd edn, 2001 Search PubMed.
- F. Makihara, M. Tsuzuki, K. Sato, S. Masuda, K. V. P. Nagashima, M. Abo and A. Okubo, Arch. Microbiol., 2005, 184, 56–65 CrossRef CAS PubMed.
- J. P. M. Jore1, N. van Luijk, R. G. M. Luiten, M. J. van der Werf and P. H. Pouwels, Appl. Environ. Microbiol., 2001, 67, 499–503 CrossRef PubMed.
- L. Jiang, J. F. Wang, S. Z. Liang, X. N. Wang, P. L. Cen and Z. N. Xu, Bioresour. Technol., 2009, 100, 3403–3409 CrossRef CAS PubMed.
- K. A. De Smet, A. Weston, I. N. Brown, D. B. Young and B. D. Robertson, Microbiology, 2000, 146, 199–208 CAS.
- L. Q. Zhu, P. L. Wei, J. Cai, X. C. Zhu, Z. M. Wang, L. Huang and Z. N. Xu, Bioresour. Technol., 2012, 112, 248–253 CrossRef CAS PubMed.
- Y. F. Zhu, J. H. Li, M. Tan, L. Liu, L. L. Jiang, J. Sun, P. Lee, G. C. Du and J. Chen, Bioresour. Technol., 2010, 101, 8902–8906 CrossRef CAS PubMed.
- L. P. Parizzi, M. C. B. Grassi, L. A. Llerena, M. F. Carazzolle, V. L. Queiroz, I. Lunardi, A. F. Zeidler, P. J. P. L. Teixeira, P. Mieczkowski, J. Rincones and G. A. G. Pereira, BMC Genomics, 2012, 13, 562 CrossRef CAS PubMed.
- M. T. Alvarez, T. Pozzo and B. Mattiasson, Biotechnol. Lett., 2006, 28, 175–181 CrossRef CAS PubMed.
- L. Jiang, J. F. Wang, S. Z. Liang, J. Cai and Z. N. Xu, Appl. Biochem. Biotechnol., 2011, 165, 98–108 CrossRef CAS PubMed.
- Z. Jin and S. T. Yang, Biotechnol. Prog., 1998, 14, 457–465 CrossRef CAS PubMed.
- A. Gupta and A. K. Srivastava, Biotechnol. Bioprocess Eng., 2001, 6, 1–5 CrossRef CAS.
- J. Coral, S. G. Karp, L. P. S. Vandenberghe, J. L. Parada, A. Pandey and C. R. Soccol, Appl. Biochem. Biotechnol., 2008, 151, 333–341 CrossRef CAS PubMed.
- F. Chen, X. H. Feng, H. Xu, D. Zhang and P. K. Ouyang, J. Biotechnol., 2012, 164, 202–210 CrossRef CAS PubMed.
- J. C. Argüelles, Arch. Microbiol., 2000, 174, 217–224 CrossRef.
- A. Wolf, R. Krämer and S. Morbach, Mol. Microbiol., 2003, 49, 1119–1134 CrossRef CAS.
- E. O. Voit, J. Theor. Biol., 2003, 223, 55–78 CrossRef CAS.
- J. E. Purvis, L. P. Yomano and L. O. Ingram, Appl. Environ. Microbiol., 2005, 71, 73761–73769 CrossRef PubMed.
- A. L. Carvalho, F. S. Cardoso, A. Bohn, A. R. Neves and H. Santos, Appl. Environ. Microbiol., 2011, 77, 4189–4199 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4gc01256a |
| This journal is © The Royal Society of Chemistry 2015 |