Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Selected dietary (poly)phenols inhibit periodontal pathogen growth and biofilm formation
Muhammad
Shahzad
a,
Emma
Millhouse
b,
Shauna
Culshaw
b,
Christine A.
Edwards
a,
Gordon
Ramage
b and
Emilie
Combet
*a
aHuman Nutrition, School of Medicine, Glasgow Royal Infirmary, College of Medical, Veterinary and Life Sciences, The University of Glasgow, Glasgow G31 2ER, UK. E-mail: Emilie.CombetAspray@glasgow.ac.uk; Tel: +44 (0) 141 201 8527
bInfection and Immunity Research Group, Glasgow Dental School, College of Medical, Veterinary and Life Sciences, The University of Glasgow, Glasgow G2 3JZ, UK
First published on 24th December 2014
Abstract
Periodontitis (PD) is a chronic infectious disease mediated by bacteria in the oral cavity. (Poly)phenols (PPs), ubiquitous in plant foods, possess antimicrobial activities and may be useful in the prevention and management of periodontitis. The objective of this study was to test the antibacterial effects of selected PPs on periodontal pathogens, on both planktonic and biofilm modes of growth. Selected PPs (n = 48) were screened against Streptococcus mitis (S. mitis), Aggregatibacter actinomycetemcomitans (A. actinomycetemcomitans), Fusobacterium nucleatum (F. nucleatum) and Porphyromonas gingivalis (P. gingivalis). The antibacterial potential of each compound was evaluated in terms of planktonic minimum inhibitory concentration (PMIC) and planktonic minimum bactericidal concentration (PMBC) using standardized broth microdilution assays. The most active PPs were further tested for their effect on mono-species and multi-species biofilms using a colorimetric resazurin-based viability assay and scanning electron microscopy. Of the 48 PPs tested, 43 showed effective inhibition of planktonic growth of one or more test strains, of which curcumin was the most potent (PMIC range = 7.8–62.5 μg mL−1), followed by pyrogallol (PMIC range = 2.4–2500 μg mL−1), pyrocatechol (MIC range = 4.9–312.5 μg mL−1) and quercetin (PMIC range = 31.2–500 μg mL−1). At this concentration, adhesion of curcumin and quercetin to the substrate also inhibited adhesion of S. mitis, and biofilm formation and maturation. While both curcumin and quercetin were able to alter architecture of mature multi-species biofilms, only curcumin-treated biofilms displayed a significantly reduced metabolic activity. Overall, PPs possess antibacterial activities against periodontopathic bacteria in both planktonic and biofilm modes of growth. Further cellular and in vivo studies are necessary to confirm their beneficial activities and potential use in the prevention and or treatment of periodontal diseases.
Introduction
Periodontitis (PD) is a common chronic disease triggered by the bacterial biofilm of dental plaque, resulting in inflammatory loss of tooth supporting tissues.1 It is a major public health issue of significant economic impact, affecting 10–15% of the world adult population2 and is the most common cause of tooth loss in adults.3 Moreover, a number of clinical and animal studies have identified positive and independent relationships between chronic periodontitis and other systemic conditions, including diabetes, cardiovascular and respiratory diseases.4–6In humans, PD is commonly associated with a dental plaque biofilm that accumulates on the tooth surfaces. It is one of the most complex and diverse microbial ecosystems within the human body, encompassing some 700 different reported bacterial species.7 However, initiation and causation of periodontitis is associated with a few pathogenic species, including P. gingivalis and P. intermedia, and A. actinomycetemcomitans in aggressive periodontitis.8
Current management approaches include periodontal surgery, scaling and root planning along with adjunctive antibiotic therapy.9 However, these treatment options not only have limited effectiveness in high risk populations and in those with advanced periodontal disease,10 but can also be associated with adverse side effects and antibiotic resistance.11 Therefore, alternative therapeutic and preventive measures which are safe, effective and free of side effects, are highly desirable.12
In recent years, an increasing interest has been observed in the use of natural compounds (of dietary origin) for the management of oral infectious diseases, including caries,13 oral candidosis14 and periodontitis.15 While plant-derived phytochemicals, especially the (poly)phenols (PPs), have been studied for their antioxidant and anti-inflammatory properties,16 the antimicrobial properties of PPs have received limited exposure in the context of prevention and treatment of PD.17 Nevertheless, several studies have reported the inhibitory activities of certain PP extracts, including cranberries, lotus, seaweed and perilla seeds against a range of planktonic periodontal pathogens.18–21 Moreover, the antibacterial activity of flavan-3-ol-rich green tea extracts has been reported against planktonic S. mitis, P. gingivalis and F. nucleatum.22–25 Others have quantified the antibacterial activity of specific PP molecules (including gallic acid, naringin and quercetin) on planktonic P. gingivalis and F. nucleatum.21,26–28 In addition, there are reports that the antimicrobial potential of PPs depends on their chemical structure.29,30 Given that periodontal pathogens in dental plaque exist in the form of biofilm, there has been very limited research investigating the effect of PPs on biofilm development and formation. Therefore, the aim of the present study was to evaluate the potential of a panel of PPs in inhibiting growth and biofilm formation of PD pathogens, with emphasis on structure–function relationship, if any.
Materials and methods
Bacterial strains, media and culture conditions
The bacterial strains used in the study were S. mitis NCTC 12261, A. actinomycetemcomitans ATCC 43718, P. gingivalis ATCC 33277 and F. nucleatum ATCC 10953. Prior to each experiment, working stocks were prepared by plating bacterial cells from frozen stocks (Microbank vials, Pro-Lab Diagnostics, Cheshire, UK). S. mitis and A. actinomycetemcomitans were maintained on Columbia agar plates with 5% defibrinated horse blood (E & O laboratories, UK; Ref DHB 100), then placed in a 5% CO2 incubator for 24 h at 37 °C. P. gingivalis and F. nucleatum were inoculated on fastidious blood agar supplemented with 5% defibrinated horse blood and incubated in an anaerobic chamber containing 85% N2, 10% CO2 and 5% H2 (Don Whitley Scientific Limited, UK) at 37 °C for 72 h. Prior to each experiment, the test strains were grown to stationary phase using appropriate nutrient broth: tryptic soy broth (Sigma, Poole, UK) supplemented with 0.8% glucose and 0.6% yeast extract for S. mitis and A. actinomycetemcomitans, and Schaedler anaerobic broth (Oxoid, Cambridge, UK) for F. nucleatum and P. gingivalis and respective growth conditions. Bacterial cultures were washed by centrifugation at 3000 rpm for 5 min in PBS and the cell count estimated using spectrophotometer, and confirmed by plate counting (CFU mL−1).(Poly)phenolic compounds
Forty eight purified (HPLC grade) PP compounds (Sigma Aldrich, Poole, UK) were used in this study. Depending on solubility, a fresh stock concentration of each PP was prepared either in water or dimethyl sulfoxide (DMSO, Sigma Aldrich, Poole, UK) and stored at −20 °C until required. Dilutions of the stock were made in respective growth media for each strain. The final concentration of DMSO was adjusted to less than 3% (v/v), which was shown to have no discernible effect on the growth of test strains (data not shown). All solution preparatory steps were carried out in a laminar flow hood (Microflow, Hampshire, England, UK).Planktonic susceptibility testing
The inhibitory potential of PPs (n = 48) against planktonic growth of the test strains (n = 4) was assessed in terms of planktonic minimum inhibitory concentration (PMIC) and planktonic minimum bactericidal concentration (PMBC) using broth microdilution technique following standard CLSI guidelines.31 Briefly, standardized (1 × 105 CFU mL−1) inoculums of the test strain were prepared in appropriate nutrient broth. The inoculum was loaded into 96-well, round bottom, polystyrene microtitre plates (Corning Incorporated, NY, USA) containing serial double dilutions of each PP (within their soluble range and triplicates). After incubation, the plates were visually inspected for any visible growth. The maximum dilution of the test compound that inhibited visible growth was recorded as PMIC, and the PMBC was determined by inoculating from those wells at concentrations ≥PMIC and assessing growth thereafter on their respective agar plate. The first concentration with no growth was recorded as the PMBC.Manufacture of hydroxyapatite coated plates
The effect of PPs on biofilm formation and maturation was assessed using flat bottom, 96-well polystyrene microtitre plates (Corning Incorporated, NY, USA), coated with hydroxyapatite (HA, Sigma Aldrich, Poole, UK; Product no 289396) by modification of the method described by Schilling et al.32 Briefly, a uniform (10% w/v) HA suspension was prepared in acetone. It was homogenized prior to transfer (60 μL) into each well of the microtitre plate. The plates were maintained on an orbital shaker set at 200 rpm and in 37 °C incubator for 15 min. After drying, loose HA powder was removed by tapping the plate, which was then washed twice in PBS and dried overnight at 37 °C. Before use, the plates were sterilized by soaking in 70% ethanol for 10 min, washed, dried and placed in an ultraviolet cabinet for 30 min.Adsorption of (poly)phenols to the substrate HA
Adsorption affinity of PPs was assessed following the method of Shellis et al.,33 with slight modifications. Briefly, 1.5 mL stock solutions (50 μg mL−1) of selected PPs (n = 10) were added into Eppendorf tubes containing 200 mg of fresh HA powder. Appropriate controls (PPs without HA) and blanks (HA and water) were also included. PPs were allowed to adsorb onto HA by incubating for 1 h at room temperature with occasional inversion and protection from light. After incubation, PP/HA suspensions were centrifuged at 5000 g for 5 min and the supernatants analysed for total phenol content using the Folin–Ciocalteau method.34 The total amount of PPs adsorbed per gram of HA (μg g−1 of HA) was determined by measuring the difference in PP concentration between sample and control. All experiments were carried out on two different occasions using samples and controls in triplicate.Effect of selected (poly)phenols adsorption to HA on adhesion of S. mitis
A viability assay based on reduction of resazurin by metabolically active cells35 was used to assess the effect of PP adsorption on HA-coated plates (biofilm substrate), prior to adhesion of S. mitis and biofilm formation. First, artificial saliva (AS) was prepared by adding porcine stomach mucin (0.25% w/v), sodium chloride (0.35 w/v), potassium chloride (0.02 w/v), calcium chloride dihydrate (0.02 w/v), yeast extract (0.2 w/v), Lab-Lemco powder; meat extract made from specially selected raw materials to enhance bacterial growth in media (0.1 w/v; Oxoid, Cambridge, UK; product code LP0029), proteose peptone (0.5 w/v) into ddH2O (Sigma, Poole, UK).36 This was sterilised by autoclaving prior to addition of urea (0.05% v/v), with a final pH of 7.4. Subsequently, AS containing PMIC concentration of selected PPs was added into each well of a HA-coated 96 well plate. Plates were sealed with adhesive lids and stored at 4 °C overnight to allow adsorption. The following day, PP solutions were removed by pipetting, and excess media allowed to evaporate in a laminar flow cabinet. Thereafter, standardized (1 × 107 cells mL−1) S. mitis suspension in AS was added to the wells. The bacteria were allowed to adhere for 4 h at 37 °C. After incubation, non-adherent cells were removed by rinsing the wells twice with PBS before adding TSB containing 10% (v/v) AlamarBlue® (Life Technologies Ltd, Paisley, UK).37 The plate was incubated for 2 h at 37 °C, after which the AlamarBlue® suspension was transferred into a clean microtitre plate, and the absorbance measured at 570 and 600 nm using a microplate reader (FluoStar Omega, BMG Labtech, UK). A percentage reduction in biofilm viability was calculated as described previously.38 The experiment was performed on two different occasions using six replicates.Determining the effect of (poly)phenols on maturation of single-species S. mitis biofilms
In order to evaluate the inhibitory effects of PPs on freshly adhered sessile cells, a standardized inoculum suspension (1 × 107 CFU mL−1) of S. mitis was prepared in artificial saliva (AS). An aliquot (200 μL) was then transferred into each well of a HA-coated 96-well plate. Following 4 h of adhesion, the media containing non-adhered cells was removed by pipetting and the biofilms washed twice with PBS. Subsequently, AS containing PMIC concentrations of selected PPs were added into each well, and further incubated for 20 h at 37 °C. Appropriate positive and negative (growth and sterility) controls were also included. Following incubation, biofilm containing wells were washed with PBS and viability of the biofilms assessed using AlamarBlue® as described in the previous section.37 The experiment was performed on two independent time points using six replicates.Effect of (poly)phenols on metabolic activity and ultra-structure changes in a mature, multi-species, pathogenic biofilm
The effect of selected PPs on the metabolic activity of a mature multi-species biofilm was assessed using an in vitro biofilm model developed by our group.38 Biofilms were formed on pre-sterilised, HA discs (Clarkson Chromatography Products Inc., Williamson, PA, USA). To produce multi-species biofilm, 500 μL of standardized (1 × 107 CFU mL−1) S. mitis (pioneer species) inoculum in AS was added to 24-well plates containing HA discs and incubated at 37 °C, in 5% CO2 for 24 h. Following incubation, the supernatant was removed and fresh AS containing standardized F. nucleatum added, which was further incubated anaerobically for 24 h at 37 °C. The supernatant was removed and standardised P. gingivalis and A. actinomycetemcomitans added. The plates were incubated anaerobically for a further four days with fresh AS replaced daily. On the seventh day, the biofilms were washed twice with PBS before addition of fresh AS containing PMIC concentration of the selected PPs. After 24 h incubation, the metabolic activity of the cells was assessed using the AlamarBlue® assay, as described above.To assess ultra-structural change of biofilms following PP treatment, scanning electron microscopy (SEM) was performed on PP-treated, mature, multispecies biofilms and untreated controls. Briefly, cells were standardised as described above, and grown directly onto Thermanox™ coverslips (Nunc, Roskilde, Denmark) to allow biofilm formation. Following maturation, biofilms were carefully washed with PBS and treated with PMIC concentrations of the selected PPs for 24 h. Following treatment, biofilms were again washed with PBS and then fixed in 2% para-formaldehyde, 2% gluteraldehyde and 0.15 M sodium cacodylate, and 0.15% w/v Alcian Blue, pH 7.4, and prepared for SEM as previously described.39 The specimens were sputter-coated with gold and viewed under a JEOL JSM-6400 scanning electron microscope. Images were assembled using Photoshop (Adobe, San Jose, CA, USA).
Statistical analysis
Data distribution and statistical analysis was performed using GraphPad Prism (Version 5; LaJolla, CA, USA). Unpaired t-tests were used to assess differences between two independent samples. Statistical significance was achieved if p < 0.05.Results
Effect of (poly)phenols on planktonic growth of test strains
We first set out to screen a panel of PPs on our test organisms to determine if any showed antimicrobial activity. An indicative selection of 48 PPs from two major groups, flavonoids (n = 27) and non-flavonoids (n = 21) were selected. Based on solubility, a range of serial double dilutions of each PP was tested, and the PMIC and PMBC determined (Table 1). All PPs tested displayed antimicrobial activity against one or more test strains, except for kaempferol-3-glucuronide, epicatechin, malvin (highest concentration tested: 250 μg ml−1), theaflavin and procyanidin B2 (highest concentration tested: 125 μg ml−1). Among all the PPs tested, curcumin was the most potent inhibitor of planktonic growth, with an MIC range of 7.81–62.5 μg mL−1, against all test strains. The highest activity was shown against P. gingivalis (PMIC = 7.81 μg mL−1) and A. actinomycetemcomitans (PMIC = 15.62 μg mL−1). Curcumin was followed by pyrogallol (very wide PMIC range; 2.4–2500 μg mL−1), pyrocatechol (PMIC range 4.88–312.5 μg mL−1) and quercetin (PMIC range 31.25–500 μg mL−1). Apigenin and daidzein were effective only against anaerobic bacteria (PMIC range 15.6–125 μg ml−1 and 500 μg ml−1, respectively) at the concentrations tested.| Range testeda | S. mitis | A. actinom. | P. gingivalis | F. nucleatum | |||||
|---|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | ||
| a Range of test concentration, All MIC values are in μg mL−1. A actinom. = A. actinomycetemcomitans. | |||||||||
| Hydroxybenzoic acids | |||||||||
| Benzoic acid | 4.88–2500 | 1250 | >2500 | 625 | 1250 | 625 | >2500 | 625 | 625 |
| 3-Hydroxybenzoic acid | 4.88–2500 | 2500 | 2500 | 625 | 1250 | 1250 | >2500 | 1250 | >2500 |
| 4-Hydroxybenzoic acid | 4.88–2500 | 1250 | 2500 | 625 | 1250 | 2500 | >2500 | 1250 | >2500 |
| 3,4-Dihydroxybenzoic acid | 4.88–2500 | 1250 | 2500 | 312.5 | 312.5 | 1250 | 2500 | 1250 | 1250 |
| Gallic acid | 4.88–2500 | 312.5 | 1250 | 9.6 | 9.6 | 1250 | 2500 | 2500 | 2500 |
| Hippuric acid | 4.88–2500 | 2500 | 2500 | 1250 | 1250 | 1250 | >2500 | 2500 | >2500 |
| Syringic acid | 1.46–750 | >750 | >750 | 750 | 750 | 375 | >750 | 750 | 1500 |
| Vanillic acid | 2.92–1500 | 750 | >1500 | 325 | 650 | 1500 | >1500 | 1500 | 1500 |
| Hydroxycinnamic acids | |||||||||
| Chlorogenic acid | 4.88–2500 | >2500 | >2500 | 312.5 | 312.5 | 312.5 | >2500 | 525 | >2500 |
| Coumaric acid | 2.92–1500 | 1500 | 1500 | 375 | 375 | 750 | >1500 | 750 | 1500 |
| Ferulic acid | 1.46–750 | >750 | >750 | 375 | 750 | 750 | >750 | 375 | >750 |
| Caffeic acid | 0.97–500 | >500 | >500 | 62.5 | 62.5 | 62.5 | >500 | 62.5 | 500 |
| Hydroxyphenylacetic acid | |||||||||
| Phenylacetic acid | 4.88–2500 | 1250 | 2500 | 625 | 1250 | 2500 | 2500 | 1250 | >2500 |
| 3-Dihydroxyphenylacetic acid | 4.88–2500 | 2500 | 2500 | 1250 | 1250 | 2500 | 2500 | 2500 | >2500 |
| 3,4-Dihydroxyphenylacetic acid | 4.88–2500 | 312.5 | 2500 | 4.88 | 4.88 | 2500 | 2500 | 2500 | 2500 |
| 3-Methoxy-4-hydroxyphenylacetic acid | 4.88–2500 | 2500 | 2500 | 1250 | 1250 | 2500 | 2500 | 2500 | >2500 |
| Flavonols | |||||||||
| Quercetin | 0.97–500 | 250 | >500 | 31.25 | 31.25 | 62.5 | >500 | 500 | >500 |
| Quercetin 3-O-glucoside | 0.488–250 | >250 | >250 | 325 | 325 | 125 | >250 | 250 | >250 |
| Quercetin 3-O-glucuronide | 0.488–250 | >250 | >250 | >250 | >250 | >250 | >250 | 250 | >250 |
| Kaempferol 3-O-rutinoside | 0.24–125 | >125 | >125 | >125 | >125 | 125 | >125 | 125 | >125 |
| Kaempferol 3-O-glucuronide | 0.488–250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 |
| Kaempferol 7-O-neohesperidoside | 0.24–125 | >125 | >125 | >125 | >125 | 125 | >125 | 125 | >125 |
| Flavanols | |||||||||
| Theaflavin extract | 0.488–250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 |
| Procyanidin B2 | 0.488–250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 |
| Catechin | 0.488–250 | >250 | >250 | 250 | 250 | 250 | >250 | >250 | >250 |
| Epicatechin | 0.488–250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 |
| Epigallocatechin | 0.488–250 | 250 | >250 | 31.25 | 31.25 | 250 | >250 | 125 | >250 |
| Epigallocatechin 3-O-gallate | 4.88–2500 | 312.5 | 625 | 39.06 | 39.06 | 156.25 | 156.25 | 312.5 | 312.5 |
| Epicatechin 3-O-gallate | 0.488–250 | >250 | >250 | 250 | 250 | 125 | 250 | >250 | >250 |
| Catechin 3-O-gallate | 0.24–125 | >250 | >250 | >250 | >250 | 125 | 125 | >250 | >250 |
| Flavanones | |||||||||
| Naringenin | 2.92–1500 | 162.5 | 325 | 162.5 | 162.5 | 46.87 | 375 | 162.5 | 325 |
| Naringin | 2.92–1500 | >1500 | >1500 | >1500 | >1500 | 750 | 1500 | 1500 | >1500 |
| Rutin hydrate | 0.97–500 | 500 | >500 | 250 | >500 | >500 | >500 | 500 | >500 |
| Hesperitin | 0.97–500 | 125 | 125 | 62.5 | 62.5 | 15.62 | >500 | 125 | 125 |
| Hesperidin | 0.97–500 | >500 | >500 | 31.25 | 31.25 | 500 | >500 | >500 | >500 |
| Anthocyanins | |||||||||
| Malvidin | 0.488–250 | >250 | >250 | 62.5 | 62.5 | 31.25 | 62.5 | 125 | >250 |
| Malvin | 0.488–250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 | >250 |
| Malvidin 3-O-galactoside | 0.488–250 | >250 | >250 | 125 | 125 | 125 | >250 | >250 | >250 |
| Pelargonidin | 0.97–500 | >500 | >500 | 62.5 | 62.5 | 62.5 | >500 | >500 | >500 |
| Oenin | 0.488–250 | >250 | >250 | 125 | 125 | >250 | >250 | >250 | >250 |
| Pelargonin | 0.488–250 | >250 | >250 | >250 | >250 | 125 | >250 | >250 | >250 |
| Flavones | |||||||||
| Apigenin | 0.488–250 | >250 | >250 | >250 | >250 | 15.62 | >250 | 125 | >250 |
| Isoflavonoids | |||||||||
| Daidzein | 0.488–250 | >250 | >250 | >250 | >250 | 500 | >250 | 500 | >250 |
| Phenolics | |||||||||
| Pyrocatechol | 4.88–2500 | 312.5 | 312.5 | 4.88 | 4.88 | 312.5 | 312.5 | 312.5 | 312.5 |
| Resorcinol | 4.88–2500 | 2500 | 2500 | 1250 | 1250 | 1250 | 2500 | 2500 | 2500 |
| Phloroglucinol | 2.92–1500 | 1500 | >1500 | 137.50 | 137.50 | 375 | 750 | 750 | 750 |
| Pyrogallol | 4.88–2500 | 9.76 | 19.5 | 2.4 | 2.4 | 312.5 | 312.5 | 2500 | 2500 |
| Curcumin | 0.390–200 | 62.5 | >200 | 15.62 | 15.62 | 7.81 | 15.62 | 31.25 | 62.5 |
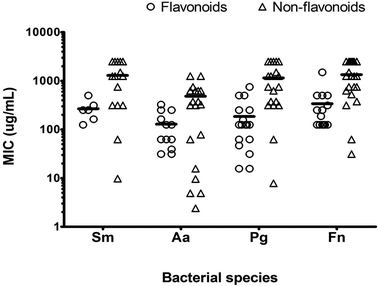
The PMBC for the majority of PPs was either the same or 2×PMIC. Wide variations in the susceptibility (mean MIC ± SD) to PPs were observed among the test strains (Table 2 and Fig. 1). A. actinomycetemcomitans showed highest susceptibility (p < 0.05) to test PPs, with a mean PMIC of 341 ± 393 μg mL−1 compared with S. mitis (1027 ± 916 μg mL−1, p < 0.001), P. gingivalis (691 ± 811 μg mL−1, p < 0.05) and F. nucleatum (941 ± 895 μg mL−1, p < 0.001) (Table 2). The order of PPs susceptibility of the test strain was A. actinomycetemcomitans > P. gingivalis > F. nucleatum > S. mitis.
| Parameter | Test strain | |||
|---|---|---|---|---|
| S. mitis | A. actinomycetemcomitans | P. gingivalis | F. nucleatum | |
| a All MIC Values are given as Mean ± S.D. (μg mL−1). Significant difference between MIC of A. actinomycetemcomitans compared with other test strains. *p < 0.05, ***p < 0.001 (T test). | ||||
| No of effective PPs | 23/48 | 35/48 | 40/48 | 35/48 |
| Range | 9.76–2500 | 2.4–1250 | 7.81–2500 | 31.25–2500 |
| MIC | 1027 ± 916*** | 341 ± 393 | 691 ± 811* | 941 ± 895*** |
Relationship between (poly)phenols structure and antibacterial activity
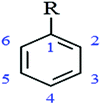
In order to assess structure–function relationship, the organism most susceptible to a broad range of PPs (A. actinomycetemcomitans) was studied.Antimicrobial activity increased considerably (lower MICs) with increasing number of OH groups attached to the benzene ring (Table 3). The antibacterial activity was also dependent on the location of the OH group. This was reflected in the higher antibacterial potential (lower PMICs) of pyrogallol (1,2,3-trihydroxybenzene; PMIC 2.4 μg mL−1) and pyrocatechol (1,2-dihydroxybenzene; PMIC 4.8 μg mL−1), having the same structure as phloroglucinol (1,3,5-trihydroxybenzene; PMIC 137.5 μg mL−1) and resorcinol (1,3-dihydroxybenzene; PMIC 1250 μg mL−1) respectively, but different hydroxylation pattern and higher PMIC values. A similar trend was observed for other PPs (Table 3). The presence of pyrogallol (3,4,5-trihydroxybenzoyl) group in the chemical structure of PPs also increased the antibacterial activity. For example, flavan-3-ols containing a pyrogallol (1,2,3-trihydroxybenzene) moiety in their structure (epigallocatechin, epigallocatechin 3-O-gallate, epicatechin 3-O-gallate) had lower PMICs (31.2, 39.0, 250 μg mL−1 respectively) and higher antibacterial activity than epicatechin and procyanidin B2 containing catechol group (1,2-dihydroxybenzene) which possessed higher PMICs (>250 μg mL−1).
| Basic structure of non-flavonoids | ||||
|---|---|---|---|---|
| PP Compound (chemical name) | Common name | Hydroxylation pattern | Substitutions R | MIC (μg mL−1) |
| 1,2-Dihydroxybenzene | Pyrocatechol | 1,2 | OH | 4.88 |
| 1,3-Dihydroxybenzene | Resorcinol | 1,3 | OH | 1250 |
| 1,2,3-Trihydroxybenzene | Pyrogallol | 1,2,3 | OH | 2.4 |
| 1,3,5-Trihydroxybenzene | Phloroglucinol | 1,3,5 | OH | 137.50 |
| 4-Hydroxybenzoic acid | 4 | COOH | 625 | |
| 3,4-Dihydroxybenzoic acid | Protocatechuic acid | 3,4 | COOH | 312.5 |
| 3,4,5-Trihydroxybenzoic acid | Gallic acid | 3,4,5 | COOH | 9.6 |
| 3-Hydroxyphenylacetic acid | 3 | CH3COOH | 1250 | |
| 3,4-Dihydroxyphenylacetic acid | Homoprotocatechuic acid | 3,4 | CH3COOH | 4.88 |
| 4-Hydroxycinnamic acid | Coumaric acid | 4 | CH–CH–COOH | 375 |
| 3,4-Dihydroxy-cinnamic acid | Caffeic acid | 3,4 | CH–CH–COOH | 62.5 |
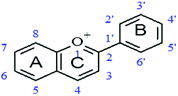
Aglycones (PP with no sugar group attached) were stronger (lower MICs) at inhibiting the growth of A. actinomycetemcomitans than corresponding glycosides (PPs with attached glucose moiety). An example is quercetin, an aglycone, which had a much lower MIC (31.25 μg mL−1) than its glycoside, quercetin-3-O-glucoside (MIC = 325 μg mL−1). The same was true for other aglycones and corresponding glycosides (Table 4).
| Basic structure of flavonoids | ||||
|---|---|---|---|---|
| PP compound (chemical name) | Common name | Hydroxylation pattern | Substitutions R | MIC (μg mL−1) |
| Quercetin 3-O-glucoside | Isoquercetin | 5,7,3,4 | 3-O-Glucoside | 325 |
| Quercetin-3-O-rutinoside | Rutin | 5,7,4′,5′ | 3-O-Rutinoside | 250 |
| 3,5,7,3′,4′-Penta-hydroxyXavone | Quercetin | 3,5,7,3′,4′ | Nil | 31.25 |
| Naringenin 7-O-neohesperidoside | Naringin | 5,7,4′ | 7-Rhamnoglucoside | >1500 |
| 5,7,4′-Trihydroxyflavanone | Naringenin | 5,7,4′ | Nil | 162.5 |
| Malvidin 3,5-O-diglucoside | Malvin | 7,4′ | 3,5-Diglucoside | >250 |
| Malvidin 3-O-glucoside | Oenin | 5,7,4′ | 3-O-Glucoside | 125 |
| Malvidin 3-O-galactoside | 5,7,4′ | 3-O-Galactocoside | 125 | |
| 3,5,7,4′-Tetrahydroxy-3′,5′-dimethoxyflavylium | Malvidin | 3,5,7,4′ | Nil | 62.5 |
| Pelargonidin 3,5-O-diglucoside | Pelargonin | 7,4′ | 3,5-Diglucoside | >250 |
| 3,5,7,4′-Tetrahydroxyflavylium | Pelargonidin | 3,5,7,4′ | Nil | 62.5 |
| Hesperetin 7-O-rutinoside | Hesperidin | 5,3′ | 7-O-Rutinoside | 62.5 |
| 5,7,3′-Trihydroxy-4′-methoxyflavanone | Hesperetin | 5,7,3′ | Nil | 31.25 |
The PMIC data was thoroughly reviewed and only a subset of the most effective PPs (those with PMIC values of <100 μg mL−1 against one or more of the test strains) were carried forward for subsequent assays. Selected PPs (n = 10) included curcumin, quercetin, EGC, EGCG, pyrogallol, gallic acid, 3,4-dihydroxyphenyl acetic acid, naringenin, hesperetin and pyrocatechol.
Effect of selected (poly)phenols on single-species S. mitis biofilms
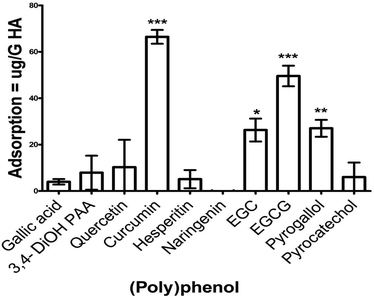
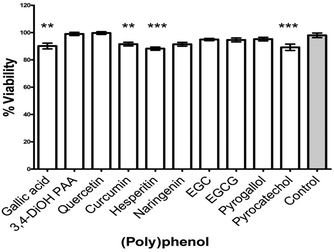
Given that S. mitis is one of the key pioneer species in dental plaque biofilm and is required for subsequent complex biofilm formation and maturation, we wanted to determine if these PPs had the capacity to inhibit different stages of S. mitis biofilm.The potential for PPs to adsorb onto HA, a key constituent of tooth enamel was tested. After 1 h incubation, the amount of PP adsorbed ranged from 1–66.5 μg g−1 HA. Compared with the control, a significant adsorption was observed for curcumin (p < 0.001), EGC (p < 0.05), EGCG (p < 0.001), and pyrogallol (p < 0.01) (Fig. 2). Other PPs (gallic acid, 3,4-dihydroxyphenyl acetic acid, naringenin, quercetin, hesperetin and pyrocatechol) did not show any significant adsorption onto HA.
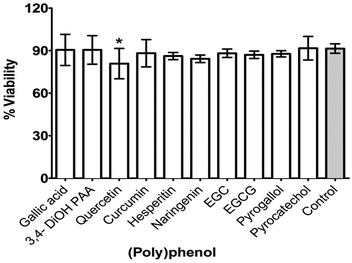
Adsorption of PP onto substrate (HA coated, 96-well plate) with the ten selected PPs (at PMIC) resulted in differential suppression of S. mitis biofilms (Fig. 3). Biofilm suppression ranged from 1 to 12% of the untreated control, with significant (p < 0.05) inhibitory activity shown by quercetin only (12%).
The ten selected PPs were also tested at their PMIC against maturation of S. mitis biofilms. Decreased viability (3–10%) in biofilms was observed following treatment with gallic acid (p < 0.01), curcumin (p < 0.001), hesperetin (p < 0.001), and pyrocatechol (p < 0.001) compared with untreated controls (Fig. 4). Other PPs (3,4-dihydroxyphenylacetic acid, quercetin, EGC, EGCG and pyrogallol) showed no significant effect on biofilm maturation.
A summary of the levels at which the ten selected PPs were effective in relation to adsorption to substrate and inhibition of biofilm formation or maturation is given in Table 5. It is interesting to note that none of the ten PPs were significantly involved in all three steps (adsorption, inhibition of formation or maturation).
| PP | Adsorption | Biofilm formation | Biofilm maturation |
|---|---|---|---|
| a 3,4-diOH PAA = 3,4-dihydroxyphenylacetic acid, EGC = epigallocatechin, EGCG = epigallocatechin 3-O-gallate. | |||
| Gallic acid | ✓ | ||
| 3,4-DiOH PAA | |||
| Quercetin | |||
| Curcumin | ✓ | ✓ | |
| Hesperitin | ✓ | ||
| Naringenin | |||
| EGC | ✓ | ✓ | |
| EGCG | ✓ | ||
| Pyrogallol | ✓ | ||
| Pyrocatechol | ✓ | ||
Bactericidal effect of curcumin and quercetin on mature, multi-species, pathogenic biofilm and ultrastructure changes
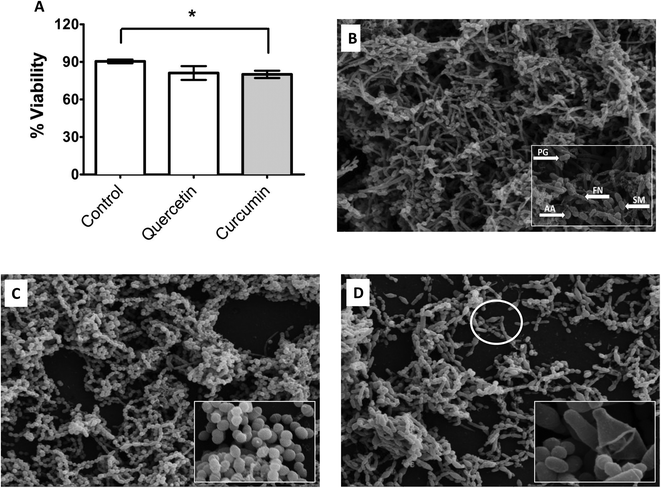
The metabolic activity of complex 4-species biofilms was significantly decreased (p < 0.05) in curcumin-treated biofilms compared with controls, but not quercetin-treated biofilms (Fig. 5A). SEM analysis of mature, multi-species biofilms treated with curcumin and quercetin are shown in Fig. 5B–D. Untreated biofilms covered the entire coverslip with all strains (S. mitis, A. actinomycetemcomitans, P. gingivalis & F. nucleatum) clearly visible and embedded in extracellular matrix (Fig. 5B), whereas curcumin-treated biofilms predominantly contain S. mitis (Fig. 5C) with no extracellular matrix visible. Biofilms treated with quercetin showed loss of biomass compared with untreated controls, and lack of F. nucleatum and extracellular matrix (Fig. 5D). Bacteria, especially P. gingivalis, appeared to swell and rupture (Fig. 5D, inset).Discussion
Global prevalence of PD, coupled with partial effectiveness of current treatments (due to development of antibiotic resistance), poses a significant challenge to scientists and oral health professionals across the world.40,41 There is, therefore, an opportunity for identification and testing of natural, alternative, therapeutic agents for prevention and/or management of the disease. Here, we have reported the antibacterial potential of PPs against key periodontal pathogens, which may be helpful in the prevention and possibly management of PD.An important consideration for cell-based assays involving PPs is their solubility. Many PPs are water insoluble and solvents such as dimethyl sulfoxide (DMSO), methanol or ethanol are often used. However, the antibacterial activity of these solvents can often interfere with the interpretation of the data.42 Keeping these limitations in mind, we first confirmed that DMSO in the concentration of 2.5% (v/v) was the most suitable solvent, having no discernible effect on the growth of the test strain (data not shown). Previously reported PMICs of PPs against P. gingivalis were noticeably different from our data (Table 1), i.e. gallic acid (MIC = 1 mg mL−1), quercetin (250 mg mL−1), naringin (250 mg mL−1), EGCG (500 μg mL−1) and EGC (1000 μg mL−1).21,23,26–28 This is, most probably, due to variations in the methods used for susceptibility testing, some authors have used the agar dilution method, different test strains, and sources of PP (commercial or natural).
A species-dependent susceptibility of periodontal pathogens to PPs was observed, where the PMIC for Gram negative test strains (A. actinomycetemcomitans, P. gingivalis & F. nucleatum) was lower than for the Gram positive test strain S. mitis (Table 2). Similar results have been obtained by Bakri and Douglas43 where Gram negative oral bacteria (A. actinomycetemcomitans, P. gingivalis & F. nucleatum) were more susceptible to the action of allicin (present in garlic extract) than streptococci. This differential activity of PPs was further reflected in the time kill curves performed on Gram positive S. mutans and Gram negative P. gingivalis.43 Among Gram-negative bacterial strains tested in our study, F. nucleatum was the least susceptible to the action of PPs and in some cases the PMIC value was approaching that for the Gram-positive strain (S. mitis). Interestingly, in addition to an outer membrane, F. nucleatum has a large periplasmic space surrounded by peptidoglycan layers, in-between the outer and inner cytoplasmic membranes.44 Therefore, it is reasonable to speculate that decreased sensitivity of the bacterium could be partly due to its membrane structure, which could hamper access of PPs to the target enzymes in cytoplasmic and perioplasmic space. Similarly, the diminished sensitivity of S. mitis may be explained by the presence of a thick peptidoglycan layer in the cell wall of Gram-positive bacteria.45 The difference between PMIC and PMBC values reflect that the PPs tested in this study were bactericidal in their activity against the test strains. These result were in agreement with a recently published study.46
Our study also reported for the first time the relationship between the chemical structure of PPs and antibacterial activity against A. actinomycetemcomitans, the most susceptible of the test strains. We have shown that increased hydroxylation of the benzene ring in phenolic acids and benzene alcohols is associated with increases antibacterial activity. The opposite association was reported previously for E. coli.29 This difference could be due to the use of different antimicrobial susceptibility assay, in addition to test strains (E. coli) and growth conditions. We showed that PPs containing a 3,4,5-trihydroxyphenyl (pyrogallol) group in their chemical structure were more potent inhibitors (possessing lower PMICs) of the growth of A. actinomycetemcomitans than PPs containing 1,2 dihydroxyphenyl (pyrocatechol) groups. These findings are in agreement with Taguri et al.47,48 who report an increased antibacterial potential of PPs containing a pyrogallol moiety against a number of food-borne pathogenic bacteria (n = 96) including both Gram positive and Gram negative strains. We also observed a stronger antibacterial activity of aglycones than their corresponding PP glycosides. PPs present in foods are mainly found in the glycosylated form however, reports indicate that PP glycosides can be hydrolysed into aglycones by salivary enzymes and the oral microflora.49,50 This further raises the possibility of increased antimicrobial activity of PPs in the oral cavity following dietary consumption that can also contribute to the prevention of oral infectious diseases including periodontitis.
The primary aetiological factor in PD is biofilm formation and its dynamic complexity. Theoretically, inhibition of the early steps in this process can result in the prevention of periodontal diseases, as has been the focus in developing vaccines against the pioneer species.51 An important property of PPs is their ability to adhere to oral hard and soft tissues, microbes and salivary proteins.52,53 Our study has confirmed that PPs, especially curcumin, possess high adsorptive affinity for HA that can be employed in increasing the local (oral) availability of PPs. This can further help in preventing and inhibiting biofilm formation on tooth surface, either through physical inhibition54 or high antimicrobial properties observed in this study and elsewhere.55 Both curcumin and quercetin can also affect the metabolic activity and architecture of mature, multi-species, pathogenic biofilm. The activity of both curcumin and quercetin appeared to be selective based on imaging of the biofilms; depleting only specific pathogens in mature biofilms, leaving a biofilm composed mainly S. mitis. This finding is interesting because S. mitis is a part of the normal oral flora, and biofilms containing only streptococci are inert and pose no threat per se to oral health. Also, swelling and rupture of P. gingivalis cells following treatment with quercetin may be a mechanism for the observed killing activity by quercetin.
Along with key strengths (the large screening, use of published guidelines, use of planktonic and biofilm assays, and single and multispecies models), this study presents some limitations. Adsorption of PPs onto HA-coated 96-well plates or commercial HA discs might be different than adsorption onto HA powder; due to differences in the available surface area and details (HA discs are more polished). Quantification of PPs adsorption onto HA discs using an ellipsometer would be helpful in this regards, and further studies combining HA with simulated salivary pellicle as a more complex surface are warranted. In addition, the antimicrobial susceptibilities and relationship between PPs structure and antimicrobial activity need to be further evaluated by using other species and clinical strains. Multispecies biofilm models have been extensively used in our group;38 their construction however can differ, and ours is completely different (both in term of the number of bacterial species and the sequence of addition of bacterial strains to produce a biofilm) from other recent work56 reporting death of S. mitis in biofilm following addition of P. gingivalis.
Collectively our observations provide evidence that the PPs tested in this study may be important in preventing oral microbial diseases and maintaining oral health. Potential routes for exploitation of PPs beneficial activities (antibacterial potential) include dietary intervention and pharmaceutical preparations. For the most active PPs, the PMICs were less than 100 μg mL−1 which can be achieved through diet as they are present in commonly consumed foods. For example, tea (green, black, oolong), turmeric and onions are rich sources of curcumin, EGCG and quercetin.57 It is estimated that one cup of tea (240 mL) contain 240 mg of EGCG.58 Moreover, the estimated daily intake of turmeric in Nepal and India is approximately 1500–2500 mg per day corresponding to 50–100 mg of curcumin.59 In the Dutch population, the estimated flavonoid intake, of which 70% is quercetin, is about 34 mg per day.60 Oral availability of PP can be potentiated by their ability to adsorb onto the tooth surface.
Where adequate intake cannot be achieved by diet alone, there are opportunities to use PPs for pharmaceutical development of oral healthcare products. Indeed, PPs containing toothpaste and dental gel are available in the market (AO ProToothpaste, AO ProVantage dental gel and Antioxidant Oral Care System marketed by PERIOSCIENCES©) containing natural (poly)phenols (phloretin & ferrulic acid) with no clinical data so far supporting a PP-specific effect on oral health. A key consideration for oral healthcare is the potential for tooth staining, which was demonstrated for tea polyphenolic extracts.33,61 Preparation of synthetic analogues with superior or optimal therapeutic properties without staining risk may be more suitable.62 We have demonstrated the antibacterial potential of PPS in vitro, further cellular and in vivo studies are required for successful use of PPs in the prevention and treatment of periodontal diseases.
References
- T. E. Van Dyke and A. J. v. Winkelhoff, J. Periodontol., 2013, 84, S1–S7 CrossRef PubMed.
- P. E. Petersen and H. Ogawa, J. Periodontol., 2005, 76, 2187–2193 CrossRef PubMed.
- P. N. Papapanou, J. Int. Acad. Periodontol., 1999, 1, 110–116 CAS.
- L. Casanova, F. J. Hughes and P. M. Preshaw, Br. Dent. J., 2014, 217, 433–437 CrossRef CAS PubMed.
- T. E. Van Dyke and J. R. Starr, Unraveling the link between periodontitis and cardiovascular disease, in J Am Heart Assoc., England, 2013, p. e000657 Search PubMed.
- X. T. Zeng, M. L. Tu, D. Y. Liu, D. Zheng, J. Zhang and W. Leng, PLoS One, 2012, 7, e46508 CAS.
- B. J. Paster, S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe and F. E. Dewhirst, J. Bacteriol., 2001, 183, 3770–3783 CrossRef CAS PubMed.
- R. Teles, F. Teles, J. Frias-Lopez, B. Paster and A. Haffajee, Periodontol. 2000, 2013, 62, 95–162 CrossRef PubMed.
- J. Slots and M. Ting, Periodontol. 2000, 2002, 28, 106–176 CrossRef.
- M. S. Tonetti, I. L. C. Chapple and W. Working Grp 3 Seventh European, J. Clin. Periodontol., 2011, 38, 114–118 CrossRef PubMed.
- P. Bidault, F. Chandad and D. Grenier, J. Can. Dent. Assoc., 2007, 73, 721–725 Search PubMed.
- E. A. Palombo, Evidence-Based Complementary and Alternative Medicine, 2011, 1–15 CrossRef PubMed.
- S. Duarte, S. Gregoire, A. P. Singh, N. Vorsa, K. Schaich, W. H. Bowen and H. Koo, FEMS Microbiol. Lett., 2006, 257, 50–56 CrossRef CAS PubMed.
- M. Shahzad, L. Sherry, R. Rajendran, C. A. Edwards, E. Combet and G. Ramage, Int. J. Antimicrob. Agents, 2014, 44(3), 269–273 CrossRef CAS PubMed.
- R. S. Percival, D. A. Devine, M. S. Duggal, S. Chartron and P. D. Marsh, Eur. J. Oral Sci., 2006, 114, 343–348 CrossRef CAS PubMed.
- S. Petti and C. Scully, J. Dent., 2009, 37, 413–423 CrossRef CAS PubMed.
- I. Palaska, E. Papathanasiou and T. C. Theoharides, Eur. J. Pharmacol., 2013, 720, 77–83 CrossRef CAS PubMed.
- V. D. La, A. B. Howell and D. Grenier, Antimicrob. Agents Chemother., 2010, 54, 1778–1784 CrossRef CAS PubMed.
- M. Li and Z. Xu, Arch. Pharm. Res., 2008, 31, 640–644 CrossRef CAS PubMed.
- J.-S. Choi, Y.-M. Ha, C.-U. Joo, K. K. Cho, S.-J. Kim and I. S. Choi, J. Environ. Biol., 2012, 33, 115–121 Search PubMed.
- H. Yamamoto and T. Ogawa, Biosci. Biotechnol. Biochem., 2002, 66, 921–924 CrossRef CAS PubMed.
- Y.-S. Cho, J. J. Oh and K.-H. Oh, Biotechnol. Bioprocess Eng., 2010, 15, 359–364 CrossRef CAS.
- S. Sakanaka, M. Aizawa, M. Kim and T. Yamamoto, Biosci. Biotechnol. Biochem., 1996, 60, 745–749 CrossRef CAS.
- M. Hirasawa, K. Takada, M. Makimura and S. Otake, J. Periodontal Res., 2002, 37, 433–438 CrossRef CAS.
- L. F. L. Elerson Gaetti Jardim Jr., A. C. Okamoto and H. Akeshigue, Arch. Health Invest., 2013, 2, 3–9 Search PubMed.
- M.-S. Kang, J.-S. Oh, I.-C. Kang, S.-J. Hong and C.-H. Choi, J. Microbiol., 2008, 46, 744–750 CrossRef CAS PubMed.
- V. W. K. Tsui, R. W. K. Wong and A. B. M. Rabie, Phytother. Res., 2008, 22, 401–406 CrossRef CAS PubMed.
- F. Geoghegan, R. W. Wong and A. B. Rabie, Phytother. Res., 2010, 24, 817–820 CAS.
- C. Cueva, M. V. Moreno-Arribas, P. J. Martin-Alvarez, G. Bills, M. F. Vicente, A. Basilio, C. L. Rivas, T. Requena, J. M. Rodriguez and B. Bartolome, Res. Microbiol., 2010, 161, 372–382 CrossRef CAS PubMed.
- A. F. Sanchez-Maldonado, A. Schieber and M. G. Ganzle, J. Appl. Microbiol., 2011, 111, 1176–1184 CrossRef CAS PubMed.
- CLSI, Methods for antimicrobial susceptibility testing of anaerobic bacteria; Approved Standard, in NCCLS Document M11-A6, Clinical and laboratory standard institute Wayne:CLSI, 2004 Search PubMed.
- K. M. Schilling, R. G. Carson, C. A. Bosko, G. D. Golikeri, A. Bruinooge, K. Hoyberg, A. M. Waller and N. P. Hughes, Colloids Surf., B, 1994, 3, 31–38 CrossRef CAS.
- R. P. Shellis, M. Addy and G. D. Rees, J. Dent., 2005, 33, 313–324 CrossRef CAS PubMed.
- V. L. Singleton, R. Orthofer and R. M. Lamuela-Raventos, Oxidants and Antioxidants, Pt A, 1999, 299, 152–178 CAS.
- E. Peeters, H. J. Nelis and T. Coenye, J. Microbiol. Methods, 2008, 72, 157–165 CrossRef CAS PubMed.
- J. Pratten, A. W. Smith and M. Wilson, J. Antimicrob. Chemother., 1998, 42, 453–459 CrossRef CAS PubMed.
- R. K. Pettit, C. A. Weber, M. J. Kean, H. Hoffmann, G. R. Pettit, R. Tan, K. S. Franks and M. L. Horton, Antimicrob. Agents Chemother., 2005, 49, 2612–2617 CrossRef CAS PubMed.
- E. Millhouse, A. Jose, L. Sherry, D. F. Lappin, N. Patel, A. M. Middleton, J. Pratten, S. Culshaw and G. Ramage, BMC Oral Health, 2014, 14, 80 CrossRef PubMed.
- S. L. Erlandsen, C. J. Kristich, G. M. Dunny and C. L. Wells, J. Histochem. Cytochem., 2004, 52, 1427–1435 CrossRef CAS PubMed.
- T. E. Rams, J. E. Degener and A. J. van Winkelhoff, J. Periodontol., 2014, 85, 160–169 CrossRef PubMed.
- C. M. Ardila, M. I. Granada and I. C. Guzman, J. Periodontal. Res., 2010, 45, 557–563 CAS.
- T. Wadhwani, K. Desai, D. Patel, D. Lawani, P. Bahaley, P. Joshi and V. Kothari, Effect of various solvents on bacterial growth in context of determining MIC of various antimicrobials, Int. J. Microbiol., 2009, 7(1) Search PubMed.
- I. M. Bakri and C. W. I. Douglas, Arch. Oral Biol., 2005, 50, 645–651 CrossRef CAS PubMed.
- A. I. Bolstad, H. B. Jensen and V. Bakken, Clin. Microbiol. Rev., 1996, 9, 55–71 CAS.
- N. Bergstrom, P. E. Jansson, M. Kilian and U. B. Skov Sorensen, Eur. J. Biochem., 2000, 267, 7147–7157 CrossRef CAS PubMed.
- Y. Asahi, Y. Noiri, J. Miura, H. Maezono, M. Yamaguchi, R. Yamamoto, H. Azakami, M. Hayashi and S. Ebisu, J. Appl. Microbiol., 2014, 116, 1164–1171 CrossRef CAS PubMed.
- T. Taguri, T. Tanaka and I. Kouno, Biol. Pharm. Bull., 2004, 27, 1965–1969 CAS.
- T. Taguri, T. Tanaka and I. Kouno, Biol. Pharm. Bull., 2006, 29, 2226–2235 CAS.
- T. Walle, A. M. Browning, L. L. Steed, S. G. Reed and U. K. Walle, J. Nutr., 2005, 135, 48–52 CAS.
- K. Kahle, M. Kempf, P. Schreier, W. Scheppach, D. Schrenk, T. Kautenburger, D. Hecker, W. Huemmer, M. Ackermann and E. Richling, Eur. J. Nutr., 2011, 50, 507–522 CrossRef CAS PubMed.
- S. Zhang, Curr. Pharm. Biotechnol., 2014, 14, 960–966 Search PubMed.
- I. Ginsburg, E. Koren, M. Shalish, J. Kanner and R. Kohen, Arch. Oral Biol., 2012, 57, 1327–1334 CrossRef CAS PubMed.
- C. J. Pan, Z. Y. Shao, J. J. Tang, J. Wang and N. Huang, J. Biomed. Mater. Res., Part A, 2007, 82, 740–746 CrossRef CAS PubMed.
- H. Ikigai, T. Nakae, Y. Hara and T. Shimamura, Biochim. Biophys. Acta, 1993, 1147, 132–136 CrossRef CAS.
- M. Daglia, Curr. Opin. Biotechnol., 2012, 23, 174–181 CrossRef CAS PubMed.
- A. E. Duran-Pinedo, V. D. Baker and J. Frias-Lopez, Infect. Immun., 2014, 82, 3374–3382 CrossRef CAS PubMed.
- J. A. Rothwell, M. Urpi-Sarda, M. Boto-Ordonez, C. Knox, R. Llorach, R. Eisner, J. Cruz, V. Neveu, D. Wishart, C. Manach, C. Andres-Lacueva and A. Scalbert, Database (Oxford), 2012, 2012, bas031 Search PubMed.
- H. Mukhtar and N. Ahmad, Am. J. Clin. Nutr., 2000, 71, 1698S–1702S CAS ; discussion 1703S–4S.
- P. Basnet and N. Skalko-Basnet, Molecules, 2011, 16, 4567–4598 CrossRef CAS PubMed.
- J. H. de Vries, P. L. Janssen, P. C. Hollman, W. A. van Staveren and M. B. Katan, Cancer Lett., 1997, 114, 141–144 CrossRef CAS.
- A. L. Cheng, C. H. Hsu, J. K. Lin, M. M. Hsu, Y. F. Ho, T. S. Shen, J. Y. Ko, J. T. Lin, B. R. Lin, W. Ming-Shiang, H. S. Yu, S. H. Jee, G. S. Chen, T. M. Chen, C. A. Chen, M. K. Lai, Y. S. Pu, M. H. Pan, Y. J. Wang, C. C. Tsai and C. Y. Hsieh, Anticancer Res., 2001, 21, 2895–2900 CAS.
- C. A. Mosley, D. C. Liotta and J. P. Snyder, Adv. Exp. Med. Biol., 2007, 595, 77–103 CrossRef.
| This journal is © The Royal Society of Chemistry 2015 |