Combined biological and abiotic reactions with iron and Fe(III)-reducing microorganisms for remediation of explosives and insensitive munitions (IM)
Jolanta B.
Niedźwiecka
and
Kevin T.
Finneran
*
Environmental Engineering and Earth Sciences, Clemson University, Biosystems Research Complex and L.G. Rich Laboratory, Clemson, SC 29634, USA. E-mail: ktf@clemson.edu
First published on 12th December 2014
Abstract
Military explosives and insensitive munitions (IM) are a significant hazard to all natural and engineered environments. A number of remediation alternatives have been proposed and investigated in detail, and biological remediation has received a large amount of attention because the reactions can be cost effective and transform the contaminants to innocuous end products. Recently, investigators have begun to look at coupled biological and chemical reactions as way to accelerate the rate of explosives and IM transformation in groundwater, surface water, soil, and industrial wastewater. A number of these technologies are predicated on reactions involving iron, and the microorganisms that reduce ferric iron. These mixed “biological–chemical” reactions may be more effective at degrading the contaminants because the strategies do not rely on a single type of reaction or microorganism, but rather a suite of reactions taking place simultaneously or in series and microbial communities that catalyze similar reactions. The technologies investigated have primarily two mechanisms. The first is initially stimulating Fe(III)-reducing microbial activity, with the secondary chemical reactions with ferrous iron (or electron shuttling molecules) actually degrading the explosives or IM. The second approach is initial chemical treatment with zero valent iron followed by engineered or native microbial activity degrading the residual contamination. In both cases the chemistry and biology of the system and the specific contaminants are critical to environmental restoration. This Frontier Review will present recent advances in the field.
Water impactCombining biological and chemical reactions can increase degradation rates for both traditional military explosives and next-generation insensitive munitions. Many bioremediation strategies require specific cells with specialized metabolic pathways for contaminant transformation. Combining biological and abiotic reactions often promotes microbial activity for populations that are less specialized and more widely distributed, which increases the number of environments in which the technology can be deployed. Remediation scientists and engineers have developed a number of systems that combine microbial and chemical processes for groundwater, soil, surface water, and industrial wastewater. |
Introduction
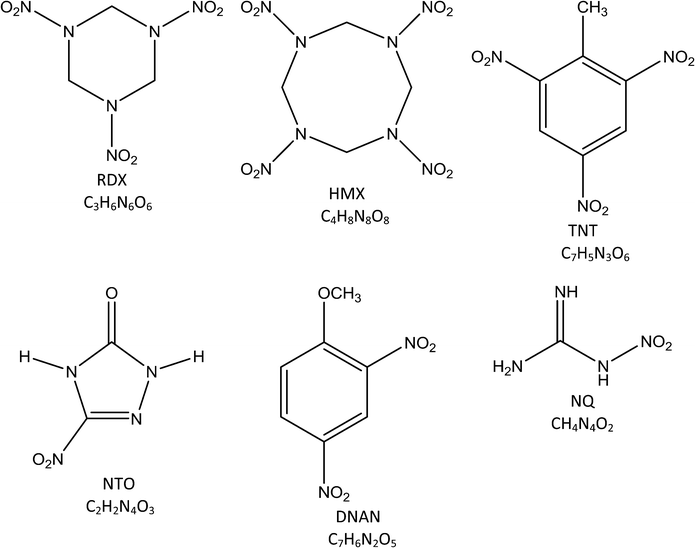
Military munitions are significant groundwater and industrial wastewater contaminants in both the United States and European Union. The compounds within munitions that are of greatest concern include the traditional nitrated explosives such as hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine (HMX), and 2,4,6-trinitrotoluene (TNT), as well as the next generation insensitive munitions (IM) such as 2,4-dinitroanisole (DNAN), nitroguanidine (NQ), and 3-nitro-1,2,4-triazol-5-one (NTO)1–3 (Table 1). These compounds are toxic to humans and wildlife; therefore, extensive research has focused on investigating their fate and transport in subsurface environments as well as treatment processes in industrial wastewater.A number of in situ and ex situ remediation strategies have been applied to explosives, and are currently being investigated for IM compounds. Biological remediation has been reported as an efficient and cost-effective approach, with long-term attenuation benefits in environments that do not threaten sensitive receptors. The underlying science for explosives remediation is relatively robust, with positive results reported in both the laboratory and field.1,3–7
Many microorganisms have been reported to degrade explosives during aerobic or anaerobic metabolic processes.8,9 In some cases the compounds are degraded to access the nitrogen within the molecules, in others the reactions are operationally defined as co-metabolic reactions where the molecules are merely electron sinks during alternative respiratory or fermentative growth. No organisms have been reported that utilize RDX or HMX as sole terminal electron acceptors in which the cells conserve energy from the reactions. These direct microbial processes can be effective, but are limited to environments where these specialized microbial populations are competitive.
More recently, research has focused on combining biological and chemical reactions for faster and more complete degradation. One strategy is to promote secondary chemical reactions – facilitated by microorganisms – to degrade explosives and IM. An example is biogenic ferrous iron mediated RDX reduction.3,10 The cells do not have to directly interact with the RDX. The ferrous iron generated during Fe(III) respiration is the direct reductant for RDX or DNAN, which is a rapid reaction at circumneutral pH3 (Fig. 1). These “mixed biotic–abiotic” reactions have several advantages over direct microbial degradation pathways. First, the cells do not necessarily have to harbour specialized enzymes for reacting with the contaminant of interest (e.g. nitroreductase enzymes11). Second, they are often easier to stimulate than the specialized microbes that directly interact with the contaminants. Finally, the lack of direct interaction allows the cells to promote degradation at both high and low concentrations, which is not always possible with direct microbial interactions.
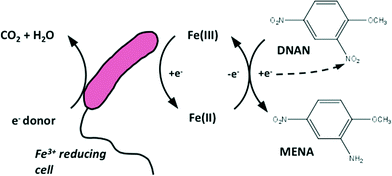 | ||
| Fig. 1 Conceptual model of mixed biotic–abiotic reactions between an Fe3+ reducing microorganism and DNAN using ferric/ferrous iron as an extracellular electron shuttling molecule. | ||
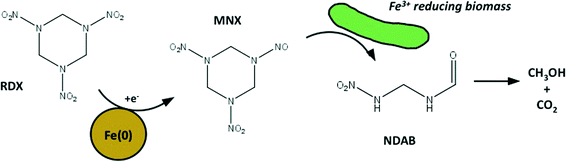
The alternate strategy is to combine chemical remediation technologies (e.g. zero valent iron or potassium permanganate4,12) with secondary biological reactions; this review will focus on zero valent iron. These coupled processes initially degrade a large fraction of the contaminant in a short period of time, while then stimulating microbial populations for long-term attenuation. They are designed to have limited operation and maintenance costs following the initial chemical remediation application. Reports suggest that these combined processes lead to different combinations of reactions products depending on the specific microbial community that develops as a result of initial treatment. Zero valent iron predominately generates nitroso intermediates from RDX (Fig. 2, first step). However, ring cleavage products resulting from the nitroso intermediates differed depending on what microbial populations were stimulated. As an example the ring-cleavage product 4-nitro-2,4-diazabutanal (NDAB) resulted when anaerobic alpha and gamma Proteobacteria were enriched during secondary microbial reactions,13 which was somewhat unusual because it was generally considered a product of aerobic microbial metabolism (Fig. 2, second step).
This frontier will describe recent developments in mixed biotic–abiotic reactions pathways for explosives and IM in groundwater and wastewater, emphasizing the new directions these technologies have taken to optimize remediation as well as strengthen the understanding of secondary reactions between microbial biomass and the contaminants of concern.
Iron and electron shuttle mediated reactions
Olivares et al. demonstrated that the insensitive munition compound DNAN was degraded under microaerophilic and strictly anoxic conditions in several soils incubated to promote microbial activity.1 DNAN was degraded to two primary intermediates: 2-methoxy-5-nitroaniline (MENA) and 2,4-diamino anisole (DAAN). While microbial activity increased the rate and extent of DNAN degradation, they reported that strictly abiotic control series also degraded DNAN while forming similar intermediates, albeit to a lesser extent. They did not identify the specific mechanism(s) for the abiotic reactions, but discussed several possibilities including ferrous iron mediated reduction, electron transfer from natural organic matter such as humic material, or sulphide mediated reduction. Subsequent reports suggest that abiotic transformation pathways facilitated by microbial activity may have contributed to the activity that was reported during stimulated biodegradation with molecular hydrogen supplemented as an electron donor. The authors then indicated caution with respect to these remediation strategies based on the formation of toxic azo and hydrazine dimmer molecules that formed as a result of all microbial and chemical reactions.Shewanella species are reported to synthesize and secrete extracellular flavin molecules to access insoluble ferric iron. Recent data demonstrated that riboflavin secreted from Shewanella putrefaciens CN32, stimulated RDX reduction in resting cell suspensions when grown with ferric iron as the primary terminal electron acceptor.14 The cells neither reduced RDX directly nor in the presence of lepidocrocite, when grown under conditions that limited or eliminated the production of the extracellular riboflavin. Conversely, when growth conditions favoured reduced riboflavin production, the cells completely removed RDX in both the presence and absence of lepidocrocite. Previous data suggested that membrane bound cytochromes are involved in RDX reduction,10 and this is an example of an alternate microbial Fe3+ reducing system that can transform RDX.
Millerick et al. reported RDX degradation for RDX mass that had been adsorbed to granular activated carbon (GAC) as a strategy to regenerate spent GAC without the need for chemical treatment or hazardous waste land filling.15 RDX was degraded directly by cells, but the rate and extent of RDX reduction was accelerated by using the electron shuttle anthraquinone-2,6-disulfonate (AQDS). The cells reduced the AQDS to anthrahydroquinone-2,6-disulfonate (AH2QDS), which chemically reduced RDX adsorbed to the surface of GAC. Formaldehyde was a transient intermediate, but it was further degraded suggesting that the coupled reactions could completely degrade RDX to innocuous end products. This was the first report of biologically mediated transformation of RDX adsorbed to GAC. The treated GAC was available for reuse, and continued to adsorb greater than 97% of aqueous RDX at starting concentrations of 100 μM with each subsequent treatment. This strategy may provide a unique platform for biologically active GAC (so called Bio-GAC) reactors, which functionally degrade RDX rather than moving from one medium (water) to another (solids).
Kwon et al. isolated and characterized a fermentative Clostridium species, C. geopurificans strain MJ1, which was capable of direct electron transfer to RDX during fermentative growth with glucose or citrate as primary fermentable substrates.7 However, the cells also reduced ferric iron and AQDS, which is similar to two other Clostridia, Clostridium strain EDB2 and Clostridium bifermentans strain HAW-1.16,17 The rate and extent of RDX degradation was accelerated in the presence of the electron shuttles for all cells, indicating that the rate-limiting step during fermentative growth is microbial reduction of extracellular molecules, and not the chemical reduction of RDX by the reduced form of the specific shuttles. The authors suggested that these types of fermenter driven secondary abiotic reactions may be a critical mechanism for RDX degradation in situ, given the prevalence of the Firmicutes during engineered remediation or monitored natural attenuation. The likely pathway for direct and indirect RDX reduction during fermentation is the hydrogen-production pathway, with the extracellular molecules replacing hydrogen production as the method by which rapidly growing fermenters dissipate excess electrons. The specific biomolecule that transfers electrons to the extracellular acceptors in these reactions has yet to be identified.
Kwon et al. also demonstrated secondary reactions with several solid phases of ferrous iron, or Fe2+ adsorbed to the surface of Fe3+ solids, during stimulated Fe3+ reduction in contaminated aquifer material from the Picatinny Arsenal in New Jersey.3 Ferrous iron adsorbed to the surface of crystalline Fe3+ was more reactive with RDX, and goethite promoted the fastest RDX reduction relative to alternate Fe3+ phases including ferrihydrite and lepidocrocite. These data agreed well with several past reports indicating that biogenic ferrous iron solids reduce RDX.18 However, these data were the first to demonstrate the relevance of this process in contaminated aquifer material. Aqueous ferrous iron alone did not reduce RDX, indicating the solid surfaces were required for catalysis. One interesting result was the microbial community that proliferated during stimulated RDX degradation. Most Fe3+ reducing environments are dominated by members of the delta Proteobacteria or the gamma Proteobacteria, with the genera Geobacter and Shewanella often comprising the majority. However, this material was overwhelmingly dominated by beta Proteobacteria (80% of all operable taxonomic units, OTUs), and of those the phylotypes most closely related to the genus Rhodoferax comprised 60% of the total OTUs. This demonstrated that the specific site geochemistry selected for unique microbial populations best suited to degrade the contaminants of concern.
Several studies in which the explosives TNT, RDX, and HMX were degraded during stimulated bioremediation in contaminated soil or aquifer material suggest that mixed biotic–abiotic reactions may be responsible for at least a fraction of the degradation activity. All of these studies employed standard electron donor amendment strategies to promote microbial activity and explosives degradation. However, the sterile controls for all materials tested transformed the contaminants, and in each case the possibility of microbially generated reduced intermediates were mentioned as possible transformation mechanisms.19,20 In the specific case of TNT transformation, the authors indicated the experimental matrix had extractable ferrous iron–ligand complexes that have been reported to reduce TNT.21 The microbial communities that developed in each case were populated with both organisms that are reported to degrade RDX/HMX/TNT directly, as well as organisms that are known to reduce Fe3+ or natural organic matter, but not the explosives. Even though understanding mixed biotic–abiotic reactions were not the goals of the studies, it was suggested in each, as one mechanism driving the reactions of interest.
Chemical treatment with secondary biological activity
Combining zero valent iron (ZVI) with secondary biological treatment is a well-developed technology. Shrout et al. demonstrated that initial ZVI treatment with electron donor stimulated microbial activity increases RDX degradation rates relative to either of the mechanisms individually.22,23 The iron was best suited for permeable reactive barrier (PRB) installations, which degraded large masses of RDX as it flowed through the reactive material. Reactions products and un-degraded RDX could then be transformed by downgradient microbial reactions, which were facilitated by electron donor amendments within or downgradient of the PRB. Ferric iron was one product of the ZVI reactions, and it became enriched downgradient of the PRB because the initial ZVI oxidation product – dissolved ferrous iron – would migrate in water away from the PRB. This Fe(III) enriched the appropriate microbial activity for continuous RDX degradation outside of the PRB. Functionally it is one of the most well optimized systems for short and long term RDX degradation. However, it is limited to groundwater plumes that are shallow and narrow enough to accommodate a PRB. Alternative approaches using ZVI (micro and/or nano scale) that was forcibly jetted into aquifer material were successful in the laboratory, but have not have the same level of in situ testing.24 This combined ZVI-microbial strategy has been recently applied to new mixtures of explosives and insensitive munitions.Ahn et al. developed an innovative treatment process to remove constituents of PAX-21 (RDX, DNAN, and ClO4−) simultaneously from wastewater.4,12 This technology was developed as an alternative to GAC adsorption combined with alkaline hydrolysis most commonly used in Army treatment plants. In the first stage, zero-valent iron (ZVI) reduced nitroaromatic and nitrated energetic compounds: RDX and DNAN. RDX was reduced to formaldehyde and DNAN was reduced to DAAN. ZVI pre-treatment rapidly removed several toxic compounds from PAX-21 wastewater that inhibited perchlorate-reducing bacteria in the anaerobic bioreactor, which was the next treatment stage. Biological perchlorate reduction is an effective strategy provided the alternate PAX-21 compounds do not interfere with the microbial activity. Formaldehyde from RDX reduction was used as electron donor for bacteria that use perchlorate as electron acceptor for cellular respiration, which negated the need for supplementing the anaerobic reactor with carbon. The combined ZVI/anaerobic bioreactor system completely removed explosives from wastewater and functioned almost as a closed loop where the products of one series of reactions fed the next series of reactions in line.
Wastewater that contains several explosives and IM combined at varying concentrations may require more than just ZVI as the reductant to catalyze the initial reactions. Koutsospyros et al. investigated the degradation of mixtures of explosives (RDX, HMX, TNT) plus insensitive-munitions (NTO, NQ, DNAN) using buffered suspensions of bimetallic particles (Fe/Cu) in an industrial wastewater stream.25 Bimetallic particles were selected in lieu of zero valent iron because of faster reaction kinetics and slower oxide formation on the particle surfaces, which delayed rust formation on the particles; rust formation led to occlusion of the active sites and subsequently loss of activity. Two types of bimetallic clusters were used: Fe/Cu and Fe/Ni. The secondary metals: Cu and Ni, increased the reduction rates by preventing oxide formation. In comparison, ZVI is strongly influenced by the formation of precipitates on the iron surface. The degradation kinetics for RDX, HMX, and TNT were similar with both bimetallic particle formulations (Fe/Cu or Fe/Ni). The insensitive compounds NQ and DNAN were degraded at rates comparable to the explosives, but NTO reduction was slower than the alternate IM tested. The primary limitation mentioned was cost, as these particles are more expensive than standard ZVI applications.
Conclusions
The number of potential applications for combined biological and chemical remediation with iron and Fe(III)-reducing microorganisms is quite large, and explosives or insensitive munitions are just two contaminant classes that can be addressed. There are a number of anecdotal data that have suggested these combined reactions were responsible for the activity reported in different environmental remediation applications.Now, scientists and engineers are designing the technologies with these mixed biotic–abiotic reactions in mind. The primary take home message to these recent data is that bioremediation scientists may have to alter the view that selecting for and stimulating a specific microbial population or community is always the correct approach. It is true that certain contaminants require the presence of specific microorganisms, such as the necessity for Dehalococcoides for complete dechlorination of trichloroethylene. However, explosives and insensitive munitions do not require specific populations, and bioremediation practitioners would be better served by strategies that are broadly applicable at a number of sites, and which do not rely on a narrow group of microorganisms. The technologies described here are just recent examples of this approach.
One possibility for future applications is directly combining chemistry and biology in single amendment technologies. As an example, porous walled microspheres are employed to reduce oxygen in radionuclide-contaminated environments via impregnated palladium catalysts. Similar delivery molecules could be developed to deliver both chemically reactive molecules (e.g. ZVI) or electron shuttling molecules, and microbial biomass to contaminated environments. With many new funding opportunities focused on broader collaborations amongst diverse teams, this is a research topic that is only in its earliest stages.
Acknowledgements
This work has been supported by the US Department of Defense Strategic Environmental Research and Development Program (SERDP) under projects ER-1377 and ER-2222. We appreciate the contribution of all past members of our research group who contributed to this work.Notes and references
- C. Olivares, J. D. Liang, L. Abrell, R. Sierra-Alvarez and J. A. Field, Biotechnol. Bioeng., 2013, 110, 1595–1604 CrossRef CAS PubMed.
- M. T. Montgomery, R. B. Coffin, T. J. Boyd and C. L. Osburn, Environ. Pollut., 2013, 174, 257–264 CrossRef CAS PubMed.
- M. J. Kwon, E. J. O'Loughlin, D. A. Antonopoulos and K. T. Finneran, Chemosphere, 2011, 84, 1223–1230 CrossRef CAS PubMed.
- S. C. Ahn, B. Hubbard, D. K. Cha and B. J. Kim, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2014, 49, 575–583 CrossRef CAS PubMed.
- F. Khan, D. Pal, A. Ghosh and S. S. Cameotra, Chemosphere, 2013, 93, 2883–2888 CrossRef CAS PubMed.
- C. H. Luo, W. O'Niell and V. Nzengung, Water, Air, Soil Pollut., 2012, 223, 5807–5815 CrossRef CAS.
- M. J. Kwon, N. Wei, K. Millerick, J. Popovic and K. Finneran, Curr. Microbiol., 2014, 68, 743–750 CrossRef CAS PubMed.
- B. Singh, J. Kaur and K. Singh, Crit. Rev. Microbiol., 2012, 38, 152–167 CrossRef CAS PubMed.
- N. N. Perreault, D. Manno, A. Halasz, S. Thiboutot, G. Ampleman and J. Hawari, Biodegradation, 2012, 23, 287–295 CrossRef CAS PubMed.
- N. N. Perreault, F. H. Crocker, K. J. Indest and J. Hawari, Can. J. Microbiol., 2012, 58, 124–131 CrossRef CAS PubMed.
- M. Roldan, E. Perez-Reinado, F. Castillo and C. Moreno-Vivian, FEMS Microbiol. Rev., 2008, 32, 474–500 CrossRef CAS PubMed.
- S. C. Ahn, D. K. Cha, B. H. Kim and S. Y. Oh, J. Hazard. Mater., 2011, 192, 909–914 CrossRef CAS PubMed.
- M. J. Kwon and K. T. Finneran, Biodegradation, 2010, 21, 923–937 CrossRef CAS PubMed.
- S. Bae, Y. Lee, M. J. Kwon and W. Lee, J. Hazard. Mater., 2014, 274, 24–31 CrossRef CAS PubMed.
- K. Millerick, S. R. Drew and K. T. Finneran, Environ. Sci. Technol., 2013, 47, 8743–8750 CAS.
- B. Bhushan, A. Halasz, S. Thiboutot, G. Ampleman and J. Hawari, Biochem. Biophys. Res. Commun., 2004, 316, 816–821 CrossRef CAS PubMed.
- H. S. Zhao, L. Paquet, A. Halasz, D. Manno and M. Hawari, FEMS Microbiol. Lett., 2004, 237, 65–72 CrossRef PubMed.
- K. B. Gregory, P. Larese-Casanova, G. F. Parkin and M. M. Scherer, Environ. Sci. Technol., 2004, 38, 1408–1414 CrossRef CAS.
- K. C. Cho, D. G. Lee, H. Roh, M. E. Fuller, P. B. Hatzinger and K. H. Chu, Environ. Pollut., 2013, 178, 350–360 CrossRef CAS PubMed.
- J. A. Livermore, Y. O. Jin, R. W. Arnseth, M. LePuil and T. E. Mattes, Environ. Sci. Technol., 2013, 47, 7672–7678 CrossRef CAS PubMed.
- N. Fahrenfeld, J. Zoeckler, M. A. Widdowson and A. Pruden, Biodegradation, 2013, 24, 179–190 CrossRef CAS PubMed.
- L. A. Sherburne, J. D. Shrout and P. J. J. Alvarez, Biodegradation, 2005, 16, 539–547 CrossRef CAS.
- J. D. Shrout, P. Larese-Casanova, M. M. Scherer and P. J. Alvarez, Environ. Technol., 2005, 26, 1115–1126 CrossRef CAS PubMed.
- G. Naja, A. Halasz, S. Thiboutot, G. Ampleman and J. Hawari, Environ. Sci. Technol., 2008, 42, 4364–4370 CrossRef CAS.
- A. Koutsospyros, J. Pavlov, J. Fawcett, D. Strickland, B. Smolinski and W. Braida, J. Hazard. Mater., 2012, 219, 75–81 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2015 |