A highly efficient designer cell for enantioselective reduction of ketones†
Gautam
Srivastava‡
,
Mohan
Pal§
,
Suneet
Kaur
and
Ravinder S.
Jolly
*
Department of Chemistry, CSIR-Institute of Microbial Technology, Sector 39, Chandigarh 160 036, India. E-mail: jolly@imtech.res.in; Fax: +91 0172 269 0585
First published on 15th September 2014
Abstract
A designer cell, surf-crs-gdh, coexpressing carbonyl reductase (crs) and glucose dehydrogenase (gdh) on the cell surface has been constructed and its enzyme activities are compared with those of the corresponding cell, cyto-crs-gdh, coexpressing crs and gdh in cytosol. For various ketones, surf-crs-gdh exhibited 48- to 265-fold higher crs activity per unit protein compared to cyto-crs-gdh.
The cofactor-dependent asymmetric reduction of ketones catalysed by alcohol dehydrogenases represents a valuable method for the synthesis of optically active alcohols.1,2 Isolated enzymes as well as whole-cell biocatalysts have been used for this purpose. However, the utility of these systems in technical applications has remained limited due to poor catalytic efficiency, especially when compared with well-established metal-catalysed asymmetric reduction.3,4 Recently, genes encoding two enzymes involved in carbonyl reduction have been co-overexpressed in suitable host cells, and applications of such “designer cells” have been demonstrated for asymmetric reduction of ketones,4 α-haloketones,5 α-hydroxy ketones,6 and α-ketoesters7 and reductive amination of α-ketoesters.8 Although these designer cells perform much better than the natural whole-cell biocatalysts in biotransformations, they still suffer from the drawback of lower efficiency due to limits imposed by the cellular membrane on substrate/cosubstrate uptake and product/coproduct efflux, which also result in complex kinetics of the overall process.9
To overcome this major drawback of designer whole-cell systems, we proposed to express these enzymes on the surface of the cell, i.e. freely hanging in the medium but firmly anchored to the outer membrane. An enzyme expressed in such a manner is expected to behave like a pure, immobilized enzyme, thereby obviating the need for cost-intensive isolation, purification and stabilization of the enzyme. Moreover, kinetics in such a system is expected to be much simpler because of the fact that substrate uptake and product efflux across the cellular membrane is not required for the reaction to occur.
The art of expressing proteins including enzymes on the surface of cells is well known and has been used in a wide range of biotechnological and industrial applications such as whole-cell biocatalysis, bioadsorbents for the removal of harmful chemicals and heavy metals, screening of human antibody libraries, mutation detection, biosensor development, etc.10
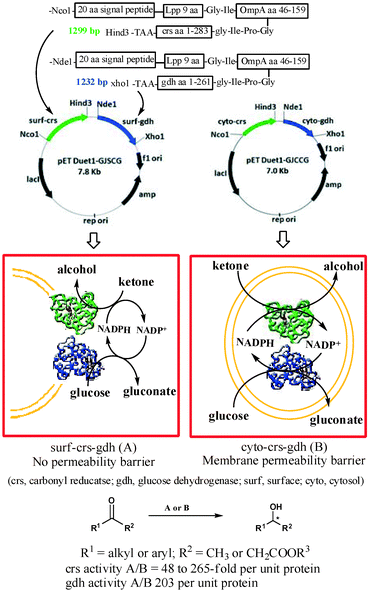
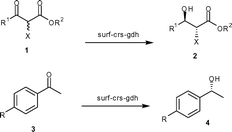
We report here a designer cell, coexpressing carbonyl reductase (crs) and glucose dehydrogenase (gdh), firmly anchored to the surface of the E. coli cell (Fig. 1). The crs activity per unit protein for the recombinant cells coexpressing crs and gdh on the surface was up to 265-fold higher compared to that of the recombinant strain coexpressing crs and gdh intracellularly. Similarly, the activity per unit protein for gdh in the recombinant E. coli strain coexpressing crs and gdh on the surface was 203-fold higher compared to that of the recombinant strain coexpressing crs and gdh intracellularly. The designer cell reduced a variety of aliphatic and aromatic ketones to furnish corresponding alcohol in 95% to >99% ee and 100% conversion.
Surface expression of crs and gdh required designing of non-natural gene sequences for each protein. The sequence designed based on literature report11 consisted of (i) an N-terminal 20-amino acid signal sequence linked to the first nine N-terminal residues of mature E. coli lipoprotein (Lpp). The 9-amino-acid residue sequence will help in anchoring the passenger protein to the outer membrane, (ii) residues 46–159 of E. coli outer membrane protein A (OmpA), which is expected to transport the passenger protein fused at its C-terminal across the membrane and (iii) a full sequence of crs (or gdh). The first 29 aa residue signal + Lpp peptide was linked to 114 aa OmpA residue through the Gly–Ile linker, which in turn was attached to the N-terminal of crs (or gdh) through Gly-Ile-Pro-Gly. The corresponding E. coli strain expressing these proteins in cytoplasm was also constructed for direct comparison of activities.12 Carbonyl reductase (crs) from Candida magnoliae was chosen as the enzyme for asymmetric reduction of ketones.13 Glucose dehydrogenase (gdh) from Bacillus megaterium was selected as the enzyme of choice for in situ cofactor recycling.14E. coli DH5α and E. coli BL21(DE3) were chosen as host cells for cloning and expression of enzymes, respectively.
Our ultimate aim was to co-express both crs and gdh together on the surface of E. coli cells. However, a priori it was not possible to predict whether or not the surface-expressed crs and gdh would adopt a native-like confirmation and would remain in active form. Therefore, as a first step we expressed crs alone on the surface of the cell to test the feasibility of the proposed study. The recombinant E. coli strain harbouring a synthetic gene for surface expression of crs has been designated as surf-crs. The corresponding strain harbouring a gene for cytoplasmic expression of crs has been designated as cyto-crs. The expression of protein in recombinant strains was confirmed by SDS-PAGE. Surface expression was confirmed by EM immunogold labelling studies (see the ESI† for details).
Because surf-crs and cyto-crs are different systems, the levels of crs-protein expressed in them may not be similar. To find the quantum of limits imposed by the cellular membrane on the efficiency of crs, it was necessary to estimate the relative amounts of crs expressed in two strains. The relative expression levels of crs were determined by an immuno-enzymatic method described in the ESI.† The surface expression level of crs was found to be 17.9-fold lower compared to the intracellular expression level of this protein. However, the recombinant E. coli strain expressing crs on the surface showed 15.7-fold higher activity for substrate 1a than the recombinant strain expressing crs intracellularly. Thus, the activity per unit crs-protein for the recombinant strain expressing crs on the surface was 275-fold higher than that for the recombinant strain expressing crs intracellularly.
The permeability of the cellular membrane is expected to vary depending on the structure of the substrate. Therefore, we tested both surf-crs and cyto-crs for reduction of a variety of ketones. The results are summarized in Table 1. As expected, surf-crs was much more efficient than cyto-crs in reduction of all of the aliphatic as well as aromatic ketones studied. The increase in activity was in the range of 50- to 275-fold per unit crs-protein.
| Ketone | surf-crs/cyto-crs | surf-crs-gdh/cyto-crs-gdh | ||
|---|---|---|---|---|
| Fold increase per unit | Fold increase per unit | |||
| Cell mass | crs-Proteina | Cell mass | crs-Proteina | |
| a crs-Protein expression per unit cell mass is 17.9-fold lower in the surf-crs strain compared to the cyto-crs strain. | ||||
| 1a | 12.72 | 275 | 14.84 | 265.5 |
| 1b | 5.37 | 96.1 | 4.84 | 86.64 |
| 1c | 3.33 | 59.6 | 3.00 | 53.70 |
| 1d | 12.25 | 219.3 | 11.01 | 197.01 |
| 1e | 5.71 | 102.2 | 5.23 | 93.62 |
| 1f | 2.86 | 51.2 | 3.04 | 54.42 |
| 3a | 3.37 | 60.3 | 3.59 | 64.25 |
| 3b | 4.88 | 87.3 | 4.56 | 81.60 |
| 3c | 5.38 | 96.3 | 5.50 | 98.45 |
| 3d | 8.96 | 160.4 | 8.16 | 146.06 |
| 3e | 2.82 | 50.5 | 2.67 | 47.79 |
| 3f | 3.41 | 61.0 | 3.07 | 54.97 |
| 3g | 10.06 | 180.1 | 9.27 | 165.93 |
| 3h | 8.06 | 144.3 | 7.42 | 132.82 |
Next, we compared the recombinant strain expressing gdh on the surface with the corresponding strain expressing gdh in cytosol. The surface expression level of gdh was found to be 13.8-fold lower compared to the intracellular expression level of these proteins (see the ESI†). However, the recombinant E. coli strain expressing gdh on the surface showed 16.3-fold higher activity than the recombinant strain expressing crs intracellularly. Thus, the activity per unit gdh-protein for the recombinant strain expressing crs on the surface was 225-fold higher than that for the recombinant strain expressing gdh intracellularly.
Finally, we constructed the recombinant strain coexpressing both crs and gdh on the surface of cells and designated it as surf-crs-gdh. The expression level and activity of enzymes in the surf-crs-gdh strain coexpressing both crs and gdh on the surface of the cell were compared with the cyto-crs-gdh strain coexpressing both crs and gdh in the cytosol of cells. The crs activity per unit crs-protein for the surf-crs-gdh strain was 265-fold higher than that for the cyto-crs-gdh strain for substrate 1a. The crs activity per unit crs-protein for substrates 1 and 3 for surf-crs-gdh was 48- to 265-fold higher than that for cyto-crs-gdh (Table 1), which is similar to that observed for surf-crs compared to cyto-crs. The gdh activity in surf-crs-gdh was about 203-fold higher per unit gdh-protein than that in cyto-crs-gdh. The enantiomeric excess and configuration of products (2 and 4) obtained from various ketones with surf-crs-gdh are shown in Table 2.
| Ketone | R 1 | R 2 | X | Yielda% | ee % | Conf. |
|---|---|---|---|---|---|---|
| a Yield of the isolated product at 100% conversion (see the ESI for reaction conditions and conversion rates). b de 99% (anti). | ||||||
| 1a | CH2Cl | CH2CH3 | H | 96 | >99 | S |
| 1b | CH3 | CH2CH3 | Cl | 91 | 98b | 2R,3R |
| 1c | CH3 | CH2CH3 | H | 89 | 95 | R |
| 1d | (CH3)CH | CH2CH3 | H | 92 | >99 | S |
| 1e | CH2Cl | n-C8H17 | H | 88 | >99 | S |
| 1f | CF3 | CH2CH3 | H | 85 | >99 | S |
| 3a | H | CH2CH3 | H | 89 | 99 | R |
| 3b | Cl | CH2CH3 | H | 92 | 99 | R |
| 3c | Br | CH2CH3 | H | 89 | 97 | R |
| 3d | F | CH2CH3 | H | 90 | 97 | R |
| 3e | CH3 | CH2CH3 | H | 85 | 99 | R |
| 3f | OCH3 | CH2CH3 | H | 87 | 98 | R |
| 3g | CF3 | CH2CH3 | H | 94 | 96 | R |
| 3h | NO2 | CH2CH3 | H | 95 | 99 | R |
An important feature from a practical point of view is that the concentration of NADPH should never become limiting for efficient conversion of ketones to alcohols. This is possible only when the enzyme responsible for recycling of the cofactor has higher activity for NADP+ to NADPH conversion than the enzyme responsible for conversion of ketone to alcohol. Gratefully, the gdh activity was about 1.9-fold higher than crs activity in the surf-crs-gdh strain co-expressing both these enzymes, which is sufficient for efficient recycling of the cofactor.
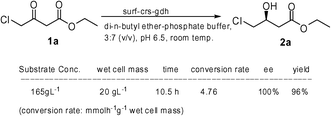
We tested the developed surf-crs-gdh biocatalyst for the production of industrially important ethyl (S)-4-chloro-3-hydroxybutyrate (2a). For industrial-scale application, it is necessary to perform the reactions at high substrate concentration. However, enzymes in general require aqueous environments in which most organic substrates are poorly soluble. Aqueous–organic biphasic systems have been successfully employed to solve this problem.12,15 We selected di-n-butyl ether as the solvent of choice after screening various short-chain ether and ester solvents. In di-n-butyl ether–aq. phosphate buffer biphasic system, the strain surf-crs-gdh at 20 g L−1 cell concentration was able to convert about 165 g L−1 (1 M) substrate 1a in 10.5 h (Scheme 1), whereas under similar conditions cyto-crs-gdh could convert a maximum of 8.25 g L−1 (0.05 M) substrate 1a.
In summary, we have shown that the recombinant E. coli strain, surf-crs-gdh, coexpressing carbonyl reductase (crs) and glucose dehydrogenase (gdh) on the surface of the cell, exhibits 48- to 265-fold higher crs activity (depending on the substrate) per unit crs-protein and 203-fold higher gdh activity per unit gdh-protein compared to corresponding E. coli coexpressing crs and gdh within (i.e. cytosol) the cells. Accordingly, the recombinant E. coli strain surf-crs-gdh may be regarded as a highly efficient designer whole-cell biocatalyst for preparation of industrially important chiral alcohols in high enantiomeric purity.
Acknowledgements
This work was supported by an NWP006 Grant from the CSIR, New Delhi, India. MP and SK acknowledge the CSIR for the award of Senior Research Fellowship. We acknowledge the technical help provided by Chander Prakash and Pradeep Patel.Notes and references
- (a) H. Gröger, W. Hummel, S. Borchert and M. Kraußer, in Enzyme Catalysis in Organic Synthesis, Wiley-VCH Verlag GmbH & Co. KGaA, 2012, pp. 1035–1110 Search PubMed; (b) K. Götz, L. Hilterhaus and A. Liese, in Enzyme Catalysis in Organic Synthesis, Wiley-VCH Verlag GmbH & Co. KGaA, 2012, pp. 1205–1223 Search PubMed; (c) K. Nakamura and T. Matsuda, in Enzyme Catalysis in Organic Synthesis, Wiley-VCH Verlag GmbH, 2008, pp. 991–1047 Search PubMed.
- M. Pal, G. Srivastava, L. S. Moon and R. S. Jolly, Bioresour. Technol., 2012, 118, 306 CrossRef CAS PubMed.
- R. Noyori and T. Ohkuma, Angew. Chem., Int. Ed., 2001, 40, 40 CrossRef CAS.
- H. Gröger, F. Chamouleau, N. Orologas, C. Rollmann, K. Drauz, W. Hummel, A. Weckbecker and O. May, Angew. Chem., Int. Ed., 2006, 45, 5677 CrossRef PubMed.
- (a) H. Gröger, F. Chamouleau, N. Orologas, C. Rollmann, K. Drauz, W. Hummel, A. Weckbecker and O. May, Angew. Chem., 2006, 118, 5806 CrossRef; (b) A. Berkessel, C. Rollmann, F. Chamouleau, S. Labs, O. May and H. Gröger, Adv. Synth. Catal., 2007, 349, 2697 CrossRef CAS.
- K. Schroer, K. Peter Luef, F. Stefan Hartner, A. Glieder and B. Pscheidt, Metab. Eng., 2010, 12, 8 CrossRef CAS PubMed.
- R. Kratzer, M. Pukl, S. Egger and B. Nidetzky, Microb. Cell Fact., 2008, 7, 37 CrossRef PubMed.
- A. Menzel, H. Werner, J. Altenbuchner and H. Gröger, Eng. Life Sci., 2004, 4, 573 CrossRef CAS.
- (a) R. R. Chen, Appl. Microbiol. Biotechnol., 2007, 74, 730 CrossRef CAS PubMed; (b) Y. Ni and R. R. Chen, Biotechnol. Prog., 2005, 21, 799 CrossRef CAS PubMed; (c) M. K. Julsing, M. Schrewe, S. Cornelissen, I. Hermann, A. Schmid and B. Buhler, Appl. Environ. Microbiol., 2012, 78, 5724 CrossRef CAS PubMed; (d) H. Nikaido, Microbiol. Mol. Biol. Rev., 2003, 67, 593 CrossRef CAS; (e) T. Nakae, Crit. Rev. Microbiol., 1986, 13, 1 CrossRef CAS PubMed; (f) L. Leive, Ann. N. Y. Acad. Sci., 1974, 235, 109 CrossRef CAS PubMed.
- For a review on cell surface display of proteins including enzymes, see: S. Y. Lee, J. H. Choi and Z. Xu, Trends Biotechnol., 2003, 21, 45 CrossRef CAS.
- (a) J. A. Francisco, C. F. Earhart and G. Georgiou, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 2713 CrossRef CAS; (b) R. D. Richins, I. Kaneva, A. Mulchandani and W. Chen, Nat. Biotechnol., 1997, 15, 984 CrossRef CAS PubMed; (c) C. Stathopoulos, G. Georgiou and C. F. Earhart, Appl. Microbiol. Biotechnol., 1996, 45, 112 CrossRef CAS.
- (a) N. Kizaki, Y. Yasohara, J. Hasegawa, M. Wada, M. Kataoka and S. Shimizu, Appl. Microbiol. Biotechnol., 2001, 55, 590 CrossRef CAS; (b) Y. Yasohara, N. Kizaki, J. Hasegawa, M. Wada, M. Kataoka and S. Shimizu, Biosci., Biotechnol., Biochem., 2000, 64, 1430 CrossRef CAS PubMed.
- M. Wada, M. Kataoka, H. Kawabata, Y. Yasohara, N. Kizaki, J. Hasegawa and S. Shimizu, Biosci., Biotechnol., Biochem., 1998, 62, 280 CrossRef CAS.
- (a) K.-D. Jany, W. Ulmer, M. Fröschle and G. Pfleiderer, FEBS Lett., 1984, 165, 6 CrossRef CAS; (b) T. Nagao, T. Mitamura, X. H. Wang, S. Negoro, T. Yomo, I. Urabe and H. Okada, J. Bacteriol., 1992, 174, 5013 CAS.
- (a) M. V. Filho, T. Stillger, M. Müller, A. Liese and C. Wandrey, Angew. Chem., Int. Ed., 2003, 42, 2993 CrossRef PubMed; (b) G. Carrea, Trends Biotechnol., 1984, 2, 102 CrossRef CAS; (c) H. GrIger, W. Hummel, S. Buchholz, K. Drauz, T. V. Nguyen, C. Rollmann, H. H. Jsken and K. Abokitse, Org. Lett., 2003, 5, 173 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Expression and immunolocalization of crs and gdh on the cell surface, experimental protocols, analytical methods, analytical data, plasmid maps, NMR spectra and gene sequence listing. See DOI: 10.1039/c4cy01017e |
| ‡ Present address: Department of Structural Biology, Weizmann Institute of Science, Rehovot 76100, Israel. |
| § Present address: Department of Biochemistry and Molecular Biology, Dalhousie University, Halifax, NS B3H 4R2, Canada. |
| This journal is © The Royal Society of Chemistry 2015 |