Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mechanical and molecular basis for the symmetrical division of the fission yeast nuclear envelope
Stefania
Castagnetti
*ab,
Bojan
Božič
c and
Saša
Svetina
*cd
aUPMC Univ Paris 06, Laboratoire de Biologie du Développement de Villefranche-sur-mer (LBDV), Observatoire Océanographique, 06230 Villefranche-sur-mer, France. E-mail: castagnetti@obs-vlfr.fr
bCNRS, Laboratoire de Biologie du Développement de Villefranche-sur-mer (LBDV), Observatoire Océanographique, 06230 Villefranche-sur-mer, France
cInstitute of Biophysics, Faculty of Medicine, University of Ljubljana, Ljubljana, Slovenia. E-mail: sasa.svetina@mf.uni-lj.si
dJožef Stefan Institute, Ljubljana, Slovenia
First published on 13th May 2015
Abstract
In fission yeast Schizosaccharomyces pombe, the nuclear envelope remains intact throughout mitosis and undergoes a series of symmetrical morphological changes when the spindle pole bodies (SPBs), embedded in the nuclear envelope, are pushed apart by elongating spindle microtubules. These symmetrical membrane shape transformations do not correspond to the shape behavior of an analogous system based on lipid vesicles. Here we report that the symmetry of the dividing fission yeast nucleus is ensured by SPB–chromosome attachments, as loss of kinetochore clustering in the vicinity of SPBs results in the formation of abnormal asymmetric shapes with long membrane tethers. We integrated these findings in a biophysical model, which explains the symmetry of the nuclear shapes on the basis of forces exerted by chromosomes clustered at SPBs on the extending nuclear envelope. Based on this analysis we conclude that the fission yeast nuclear envelope exhibits the same mechanical properties as simple lipid vesicles, but interactions with other cellular components, such as chromosomes, influence the nuclear shape during mitosis, allowing the formation of otherwise energetically unfavorable symmetrical dumbbell structures upon spindle elongation. The model allows us to explain the appearance of abnormal asymmetric shapes in fission yeast mutants with mis-segregated chromosomes as well as with altered nuclear membrane composition.
Introduction
Maintaining organelle morphology is essential for cellular function and changes in the shape of different organelles are often linked with aging, cancer and other genetic diseases. Still the mechanisms that dictate the shape of many membrane-bound organelles remain largely unknown. Membrane shape transformations have often been studied using artificial lipid vesicles, whose shapes correspond to the minimum of membrane elastic energy.1–3 However, as the plasma-membrane and membranes of cellular organelles are associated with a multitude of proteins and interact with cytoskeletal structures, the same principles may not apply when describing their shape transformations. To address this issue we analyzed experimentally and theoretically the shape transformation undergone by the nucleus during mitosis in the fission yeast Schizosaccharomyces pombe (S. pombe).The nucleus, the defining organelle of eukaryotic cells, separates the genome from the cytoplasm and is delimited by the nuclear envelope, a double lipid bilayer, composed of an inner nuclear membrane, which is in contact with the nucleoplasm and directly interacts with chromatin, and an outer nuclear membrane continuous with the endoplasmic reticulum (ER).4 The outer and inner membranes are connected at nuclear pores and thus topologically represent a single membrane. In fission yeast the nucleus is round during interphase, but it undergoes extreme shape changes during mitosis, which takes place within an intact nuclear membrane (closed mitosis).5 During metaphase and anaphase the nucleus remains spherical. As the segregated sister chromatids move away from each other in anaphase B and telophase, the nucleus elongates to accommodate the movement of the segregating chromosomes, acquiring a symmetrical dumbbell shape (Fig. 1a) before elongating and separating into two identical spherical daughter nuclei.6 Both chromosome segregation and nuclear division are driven by the mitotic spindle, a bipolar array of microtubules emanating from the spindle pole bodies (SPBs), the yeast microtubule organizing centers,7 embedded in the nuclear membrane. The mitotic spindle forms inside the nucleus during the early stages of mitosis. Polymerization and sliding of overlapping spindle microtubules, during anaphase B, result in spindle elongation, chromosome segregation and nuclear envelope division.
A possible approach to study fission yeast mitotic nuclear envelope shape transformations during microtubule spindle elongation is to consider the nuclear envelope as an elastic membrane with the same mechanical properties of a lipid membrane. Previous studies showed that microtubules growing inside a lipid vesicle cause the formation of an asymmetrical shape with a round main body and a thin tubular protrusion, known as membrane tether.8,9 Such a shape corresponds to the minimum of the system free energy, both in vesicles with a fixed surface area10,11 and those connected to a membrane reservoir.12,13 In fission yeast cells, similar asymmetric shapes with one or two tethers have been observed when microtubules extend within the nucleus without associated SPBs, both in interphase and during mitosis. Nuclear envelope protrusions have been observed in mitosis, when spindle microtubule association with SPB is impaired (acentrosomal spindle). In msd1Δ14 and cut11-215 cells, which are defective in SPB–microtubule anchoring and maintenance of the SPBs on the nuclear membrane respectively, spindle microtubules elongate in the absence of associated SPBs and the nucleus exhibits thin protrusions associated with the extending spindles. A similar nuclear protrusion phenotype is observed when anaphase B spindles are cut by laser microsurgery leaving elongating spindle fragments without associated SPBs.16 Over-expression of ned113 or mia16 causes the formation of acentrosomal intra-nuclear microtubule bundles and results in the formation of thin nuclear tethers during interphase and mitosis respectively. Taken together these observations suggested that in the absence of SPB–microtubule interactions, the nuclear envelope behaves as a true elastic entity.13 However, during an unperturbed mitosis, the fission yeast nucleus acquires a symmetrical dumbbell shape, which has never been observed when growing microtubules inside pure lipid vesicles8,9 and which, as it will be demonstrated theoretically, has higher free energy and is therefore unstable.
In fission yeast, kinetochores, the specialized multi-protein complexes assembled at centromeres to allow the interaction between mitotic chromosomes and spindle microtubules, are clustered in the vicinity of the SPB during interphase, but dissociate from the SPB upon mitotic entry to interact with spindle microtubules. Following chromosome separation, kinetochores move to spindle poles adjacent to SPBs where they remain during spindle elongation.17 Here we show that, in fission yeast, clustering of kinetochores in the vicinity of the SPB during spindle elongation is required for the formation of symmetric dumbbell nuclear shapes and demonstrate theoretically that due to such an interaction the mirror-symmetrical shapes can become energetically stable. Based on our analysis we propose that the shape behavior of the fission yeast nuclear envelope follows the same laws of simple lipid vesicles, but that other forces originating from interactions with nuclear components (SPB and chromosomes) also contribute to the energy of the system and therefore need to be considered. On the basis of the proposed model it is also possible to interpret the shape behavior of the nucleus in fission yeast mutants with mis-segregated chromosomes and with altered nuclear envelope mechanical parameters.
Experimental results
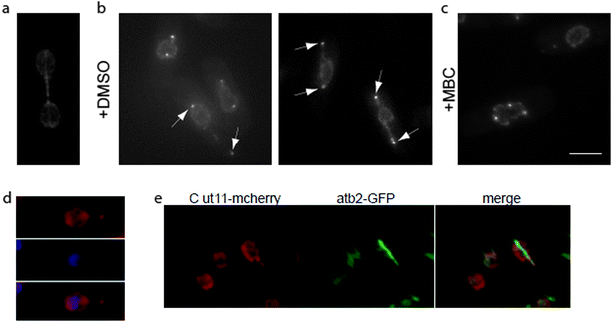
During interphase, clustering of kinetochores in the vicinity of the SPB17 is essential to minimize nuclear envelope distortion caused by cytoplasmic microtubules pulling on the SPB.18 We reasoned that SPB–kinetochore interactions during spindle elongation in anaphase B might also play a role in nuclear shape changes during mitosis, increasing the ability of the nuclear envelope to tolerate the pushing force generated by extending spindle microtubules. To test this hypothesis, we used a mutant in Nuf2, a subunit of the Ndc80 complex, a core kinetochore component.19 The temperature sensitive nuf2-3 mutant affects kinetochores clustering at SPB both in mitosis and in interphase,20,21 and it was reported that at restrictive temperature about 10% of cells block with an elongated spindle and un-segregated DNA.21 Using a cdc11-119 mutant background to avoid the formation of a septum across the undivided nucleus and Cut11-mcherry to mark the nuclear envelope, we analyzed nuclear shape changes during mitosis in cdc11-119 nuf2-3 cells. We observed the appearance of nuclear extensions in 14.5 ± 3% of cells after 3 hours at the restrictive temperature (Fig. 1b). Fission yeast cells containing nuclear tethers accumulate Cut11-mcherry on SPBs (Fig. 1b, arrowheads) at the tip of the tether. As it was previously suggested that SPBs are sufficient to maintain nuclear morphology, we confirmed the presence of SPBs at the end of nuclear tethers using the SPB marker Sad1-DsRED and observed that as for Cut11-mcherry, Sad1-DsRED was always associated with nuclear tethers (data not shown).As Cut11 accumulates on SPBs from mitotic commitment to the metaphase–anaphase transition, this result suggests that nuclear tethers form during mitosis. To confirm the cell cycle stage of these cells, we labeled microtubules using a strain bearing a GFP-tagged α-tubulin, atb2-GFP. All cells containing nuclear tethers were mitotic, as they had no cytosolic microtubules and spindle microtubules extended, inside the nucleus, along the length of the tether (Fig. 1e). Moreover, treatment with the microtubule-depolymerizing drug, carbendazim (MBC), suppressed tether formation (Fig. 1c), suggesting that their extension is a consequence of spindle microtubule elongation.
Labeling of DNA with 4,6-diamidino-2-phenylindole (DAPI, Fig. 1d) showed that DNA was absent from nuclear tethers. Taken together, these observations show that SPB association with spindle poles is not sufficient to support the changes in nuclear morphology observed during mitosis. Instead, clustering of kinetochores in the vicinity of SPBs favors the formation of spherical nuclear shapes over long tubular tethers.
Theoretical model
In the model it is assumed that as for vesicles, the energy that determines the shape of the fission yeast nucleus is the membrane bending energy.22 The integral of the area density of the bending energy over all area elements dA of the nuclear envelope with area A is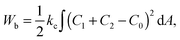 | (1) |
During closed mitosis, the nuclear area increases by 26% to allow for the transition from one to two spheres at a constant volume.13 The membrane required for nuclear growth is obtained from the ER, which is continuous with the outer nuclear membrane.23 As the membrane content of the ER is much larger than that of the nuclear envelope and phospholipid synthesis is enhanced during mitosis,24 the ER can be considered as a membrane reservoir. Taking into account the exchange of membrane between the nuclear envelope and the ER, the shape of the nuclear envelope will correspond to the minimum of the total nuclear envelope energy (W) including the bending energy described by eqn (1) and a second term corresponding to the work needed to move the membrane material from the ER to the nuclear envelope12
| W = Wb + 2σA | (2) |
The theoretical prediction for the shapes corresponding to the minimum of the energy in eqn (2) for fixed pole-to-pole distances is shown in Fig. 2. The lowest energy corresponds to an asymmetric shape composed of a lemon-like main body and a thin tubular tether. A symmetric shape with tethers on both sides of the main vesicle body has slightly higher energy, whereas, based on this model, the energy of dumbbell shapes resembling that of the dividing fission yeast nucleus is much higher and cannot be globally stable.
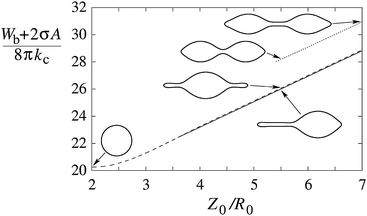 | ||
| Fig. 2 Energy of the axially strained vesicle as a function of its length, plotted for axisymmetric shapes with a single tether (full line), two tethers (dashed line) and a dumbbell shape (dotted line). The length (Z0) is given in units of R0 which is the radius of the sphere with the same volume as the volume of the nucleus, R0 = (3V/4π)1/3. Shapes are calculated on the basis of the minimization of the energy functional eqn (2) with C0 chosen as 1/R0 which would favor dumbbell shapes.3 Membrane tension is σ = 20kc/R02. | ||
We next attempted to integrate the effect of chromosome clustering at SPBs on the shape of the dividing fission yeast nucleus in our theoretical analysis. We assumed that the convexly shaped nuclear envelope at nuclear poles limits the space available to the chromosomes clustered in the vicinity of the SPBs. As a consequence, the energy, which will dictate the nuclear shape, will have two components – the bending energy of the nuclear envelope and the deformational energy of the clustered chromosomes.
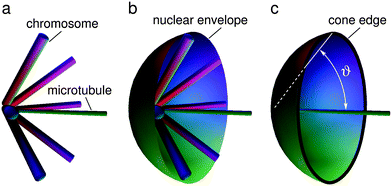
In our model the arms of condensed mitotic chromosomes are considered as elongated elastic entities that emanate from the nuclear poles (corresponding to the SPB) which would in the absence of the nuclear envelope align on the lateral surface of a cone with an azimuthal angle ϑ0 and a lateral height equal to the average length of fission yeast chromosome arms (Larm) (Fig. 3a). The elasticity of this system can be approximated by the elastic behavior of a cone whose energy increases if the azimuthal angle ϑ is either larger or smaller than its equilibrium value ϑ0 (Fig. 3). The shape of the nuclear envelope under these circumstances corresponds to the minimum of the system energy which includes the bending energy of the nuclear envelope and the deformational energy of the cones present at the two opposite sides of the nuclear envelope. In the model we took the cone deformational energy to be proportional to the product of the square of the difference ϑ − ϑ0 and the elastic constant Kϑ, Kϑ(ϑ − ϑ0)2.
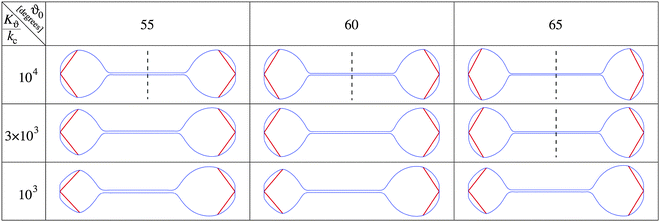
The nuclear envelope shape that corresponds to the minimum free energy of the system is sought numerically as described in the Methods. Fig. 4 shows examples of calculated shapes for a given pole to pole length at different values of equilibrium azimuthal angle ϑ0 and the ratio between the two involved elastic constants Kϑ/kc. The shapes are calculated at a constant membrane area, on the assumption that the enhanced phospholipid synthesis24 already ceased. As shown, depending on ϑ0 and Kϑ/kc, the system can be shifted to obtain both stable symmetrical or asymmetrical shapes. The symmetrical dumbbell shape observed during fission yeast mitosis can be obtained by increasing the ratio between the two elastic constants with the critical ratio being smaller at larger equilibrium angles ϑ0.
More detailed calculations of the results presented in Fig. 4 showed that the transition from symmetrical dumbbell shapes to shapes with sphere-like parts of different sizes is continuous, i.e. of the second order. Moreover, the position of this transition depends on the angle ϑ0 as well as on other system parameters. Fig. 5 shows that the range of symmetrical shapes increases at larger nuclear envelope areas and shorter microtubule lengths, and decreases at large positive or negative values of the spontaneous curvature.
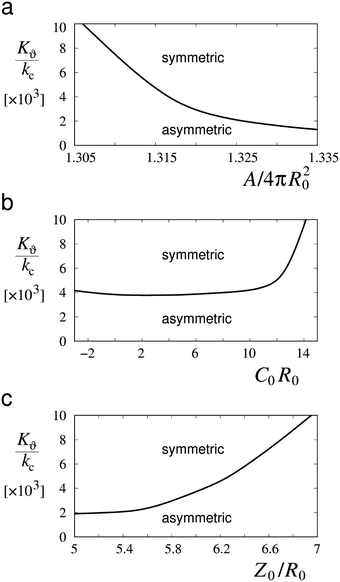 | ||
| Fig. 5 Values of the ratio between the elastic constant of chromosome arms and the bending constant of the nuclear envelope (Kϑ/kc) at which asymmetric shapes become symmetric dumbbell shapes in relation to (a) the area of the nuclear envelope (A/(4πR02)), (b) its spontaneous curvature (C0R0) and (c) the length of microtubules (Z0/R0). Plotted curves represent in all three cases the deviation from the physical parameters of the shapes in Fig. 4 at ϑ0 = 60 degrees: Z0/R0 = 6, C0R0 = 1 and A/(4πR02) = 1.317. | ||
The occurrence of symmetrical dumbbell shapes and asymmetrical shapes can be understood on the basis of the constraints that the nuclear envelope imposes on the chromosome spatial arrangements. The tendency of the nuclear envelope is to acquire an asymmetrical shape (Fig. 2) whereas the chromosomes present at opposite nuclear poles are identical and therefore apt to symmetrical deformations. If the chromosomes can be easily deformed, the tendency of the nuclear envelope prevails, and the final shape is asymmetrical. However, if small chromosome deformations require a high amount of energy, as a consequence of a relatively large value of the elastic constant Kϑ, the nuclear envelope can only respond by symmetrical distortions giving rise to a symmetrical dumbbell shape. A higher value of the ratio Kϑ/kc is required to attain a symmetrical dumbbell shape at smaller membrane areas and at longer microtubule lengths that determine the distance between the poles (Fig. 5). Variation in spontaneous curvatures between zero and six has no appreciable effect on the transition between symmetrical and asymmetrical shapes.
Discussion
This work aims at comparing the shape behavior of lipid vesicles with that of biological membranes to determine how the biomechanical characteristics of lipid membranes are modified to obtain the shape behavior of cellular organelles. Given the wealth of information available and the ease of manipulation, we chose the fission yeast nucleus as a representative example, and analyzed its shape changes during mitosis.Previous theoretical work13 has shown that during interphase, the fission yeast nucleus behaves in accordance with experimental and theoretical biomimetic studies. This analysis suggested that the nuclear envelope has the same mechanical properties of a simple lipid membrane. However, the symmetrical nuclear shapes, observed during fission yeast mitosis, cannot be described by this simple model and require the involvement of additional forces. Here we analyzed the nuclear morphology in mutants impairing kinetochore clustering at SPBs and showed that nuclear symmetry during mitosis requires SPB–chromosome interactions. Based on these observations we hypothesized that when chromosomes are not clustered in the vicinity of SPBs, they do not affect the energy of the nuclear envelope which therefore behaves like a free lipid vesicle. However, when chromosomes are associated with SPBs and maintained near the nuclear envelope at nuclear poles, the interaction between the nuclear envelope and chromosomes causes an increase in the chromosome elastic energy, which is sufficient to make the symmetrical nuclear envelope shapes energetically favorable.
As shown in Fig. 4 and 5 symmetrical divisions can occur at certain ratios of the internal energy of chromosome arms over the energy of the nuclear membrane, quantitatively expressed as the ratio between the corresponding elastic constants. Bending constants for lipid bilayers are known and it can be assumed that the bending constant of the nuclear envelope is in the order of twice the magnitude of a bilayer. Given the lack of structural information about SPB–chromosome interactions, it is necessary to make some assumptions to define the elastic deformation of chromosome arms determined by the constant Kϑ. To determine the value of Kϑ, we assumed that it depends on chromosome bending. In the corresponding model a chromosome arm is described as an elastic rod with the bending energy (Wb,chr)
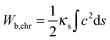 | (3) |
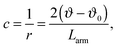 | (4) |
By inserting c from eqn (4) into eqn (3) and taking into consideration that in fission yeast there are six chromosome arms, we obtain Kϑ = 12κs/Larm. Bending constants of condensed chromosomes are in the range κs = 10−23 to 10−22 J m.26 Lipid membrane bending constants are dependent on the membrane composition in the range kc = 0.5 to 2.0 × 10−19 J.23 By taking Larm = 1 μm (average S. pombe chromosome arm length), κs = 5 × 10−23 J m, and kc = 2.5 J (taking into account that the nuclear envelope is a double layer) we obtain Kϑ/kc = 2.4 × 103 which means that in view of the result presented in Fig. 4 the establishment of symmetrical nuclear envelope shapes can be well accounted for by the elastic properties of the nuclear envelope and chromosomes.
Asymmetrical nuclear envelope division, with two nuclei of unequal size, has been observed in fission yeast mutants, which segregate their chromosome unequally to the two daughter nuclei.27 This phenotype can be easily understood based on the model suggested here. As the number of chromosomes clustered at the two SPBs is different, the chromosome–membrane interaction is also altered, causing the nuclear envelope to shift to an equilibrium corresponding to an asymmetric shape. Perhaps less intuitively, asymmetric shapes can also be obtained with equal chromosome numbers by altering the elastic properties of the membrane. Indeed, asymmetric nuclear shapes can be observed in fission yeast mutants with equal chromosome segregation, such as the lds1 (large and small daughter) mutants.28 As the lsd1 mutants affect fatty acid synthesis, it can be speculated that in these cells the nuclear envelope properties are different from wild type cells because of the altered fatty acid content. The bending constants of membranes for mixtures of phospholipid molecules with different fatty acid chains can be drastically different.29 At a constant chromosome length, a reduction in the fatty acid content causing an increase in the membrane bending constant, would result in a diminished Kϑ/kc ratio, shifting the system from the regime of wild type symmetrical nuclear envelope divisions to the observed asymmetrical divisions.
Based on our analysis we propose that the same principles apply to shape behavior of biological membranes and simple lipid vesicles. However, in a complex system such as a cell, interactions with different cellular components also contribute to the total energetic requirement, which determines membrane behavior. To understand the shape behaviors of biological membranes it is therefore important to identify by employing the appropriate cell biology methods the interactions which are critical for the performance of a specific cellular process.
Methods
Experimental
All S. pombe strains used in this study are listed in Table 1. Standard methods were used for growth and genetic manipulation.30 All experiments, unless otherwise stated, were performed in YE4S (yeast extract with added 250 mg l−1 histidine, adenine, leucine and uridine). Cells were grown at 25 °C to 1–2 × 106 cell per ml density before shifting to the restrictive temperature (36.5 °C). Cells were collected by centrifugation at hourly interval and fixed in cold 70% ethanol. Cells were re-hydrated in distilled water and mounted in 2.3 μl of 50% glycerol. For DAPI staining, the cells were mounted in 2.3 μl of 50% glycerol and 0.1 M Tris, pH 8 containing 1 μg ml−1 DAPI. For live imaging, the cells were imaged in minimal media using a Zeiss Imager A2 microscope.| Strain | Genotype | Source |
|---|---|---|
| FY15675 | h+ ura4-D18 nuf2-3::ura4+ | YGRC |
| PN113 | h+ cdc11-119 | Nurse's lab |
| PN732 | h− ura4D18 | Nurse's lab |
| SC452 | h− Heh2-GFP-kanMX ura4-D18 | Blobel's lab |
| SC482 | h+ cdc11-119 sad1DsRed:leu2 leu1-32 ura4-D18 | Castagnetti's lab |
| SC379 | h+ cdc11-119 nuf2-3:ura4 ura4-D18 | Castagnetti's lab |
| SC395 | h− cdc11-119 nuf2-3:ura4 cut11mcherry:ura4 ura4D18 | This study |
| SC449 | cdc11-119 nuf2-3:ura4 atb2-GFP-kanMX cut11-mcherry:ura4 ura4-D18 | This study |
| SC481 | cdc11-119 nuf2-3:ura4 heh2-GFP-kanMX sad1Ds-RED:leu2 leu1-32 ura4-D18 | This study |
| SC264 | cdc11-119 cut11-GFP:ura4 nmt81-atb2-GFP:kanMX ura4-D18 | Castagnetti's lab |
Theoretical
In order to obtain the minimum of the nuclear envelope energy at a given nuclear volume we chose the corresponding parameterization. As the nuclear envelope is axisymmetric along the Z-axis, the principal curvatures along the meridians and the parallels can be expressed by| Cm = dψ/dS | (5) |
Cp = sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ψ/R ψ/R | (6) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) dS, whereas the volume of the nucleus can be expressed as V = ∫πR2
dS, whereas the volume of the nucleus can be expressed as V = ∫πR2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin
sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ψdS.
ψdS.
To calculate the total nuclear envelope energy of a particular membrane shape, the contour of the membrane was expressed in a discretized representation. The continuous functions, R(S), Z(S) and ψ(S), are represented by discrete nodes [Rj = (Rj,Zj,ψj)], based on which the derivatives and integrations in all expressions are approximated. The principal curvatures can be expressed by
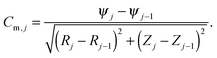 | (7) |
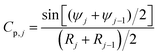 | (8) |
The procedure starts by determining the minimum of the energy for a constant membrane area. In practice, constraint-compliance energies (WV,WA) are defined. They are treated simply as fictitious energies for enforcing volume and area constraints
| WV + WA = (KV/2)(V − V0)2/V0 + (KA/2)(A − A0)2/A0 | (9) |
Wr + Wz = Λr∫(dR/dS − cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ψ)2dS + Λz∫(dZ/dS + sin ψ)2dS + Λz∫(dZ/dS + sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ψ)2dS. ψ)2dS. | (10) |
| W′ = Wb + WV + WA + Wr + Wz | (11) |
The shape of the nuclear envelope is determined iteratively in such a way that the potential W′ becomes minimal at chosen values of the parameters (V0, A0, Z0, C0 and kc). In each iteration step, the redistribution of the nodes describing the membrane shape [Rj = (Rj, Zj, ψj)] occurs. The number of nodes in the model was taken to be 159. In each step the changes in values of Rj are taken to be proportional to the corresponding derivatives of the potential, Rnewj = Roldj − h(∂W′/∂Rj), where the parameter h determines the speed of the iteration. The value of h taken is small enough to avoid the increase of the potential W′. The values of Rnewj and Znewj determine the next approximation for the nuclear envelope shape. The radius R and the position along the symmetry axis Z of the first and the last node are determined by the radius and positions of the SPBs. At the end-nodes the membrane is taken to be perpendicular to the symmetry axis. In each consecutive iteration step the corresponding corrections are generally smaller. The typical number of total iteration steps was 106–107. Usually in the initial steps the nodes were not distributed evenly, therefore their positions were regulated – if the distance between them was too large or too small, the corresponding values were changed by Rj* = (Rj+1 + xRj + Rj−1)/(x + 2) and Zj* = (Zj+1 + xZj + Zj−1)/(x + 2) with x > 2 × 103.
At a given tension in the membrane the shape cannot be directly determined. An effective way for energy minimization of W (eqn (2)) is to calculate A0 at given membrane tension (Fig. 2). Namely, A0 relates to the required tension according to the equation σ = KA(A − A0)/A0 which has to be fulfilled in the equilibrium. Thus, each iteration step is divided into two stages. In the first stage, the value of A0 is calculated from the membrane area (A) obtained in the previous step and the required membrane tension (σ). In the second stage the redistribution of the nodes occurs, according to the derivatives of the potential W′. The described method works successfully since the number of iterations is high enough.
In order to obtain the minimum of the total energy of the system, which also includes the deformation of chromosome arms, we added to the bending energy (eqn (1)) a term for the corresponding energy (Kϑ(ϑ − ϑ0)2) in the expression for W′ (eqn (11)). In this case the changes in positions of the nodes at the chromosome ends are limited to their length and they are proportional to the derivatives of the potential W′ in a plane perpendicular to the direction from the beginning to the end of the chromosomes (Fig. 6).
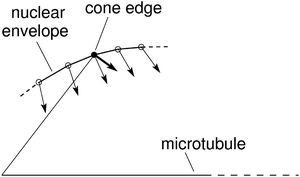 | ||
| Fig. 6 Schematic representation of node movements in the vicinity of the cone edge (see Fig. 3). The nodes that describe the nuclear envelope are denoted by small circles. The filled circle corresponds to the cone edge at the end of the chromosomes. The changes of node locations are proportional to the derivatives of W′ (thin arrows) except for the node that corresponds to the cone edge (thick arrow). | ||
Acknowledgements
The work was supported by the Slovenian Research Agency through the research program P1-0055.References
- H. J. Deuling and W. Helfrich, J. Phys., 1976, 37, 1335–1345 CAS.
- S. Svetina and B. Žekš, Eur. Biophys. J., 1989, 17, 101–111 CrossRef CAS.
- U. Seifert, K. Berndl and R. Lipowsky, Phys. Rev. A: At., Mol., Opt. Phys., 1991, 44, 1182–1202 CrossRef CAS.
- M. L. Watson, J. Biophys. Biochem. Cytol., 1955, 1, 257–270 CrossRef CAS.
- I. B. Heath, Int. Rev. Cytol., 1980, 64, 1–80 CAS.
- L. Zheng, C. Schwartz, V. Magidson, A. Khodjakov and S. Oliferenko, PLoS Biol., 2007, 5, e170 Search PubMed.
- S. L. Jaspersen and M. Winey, Annu. Rev. Cell Dev. Biol., 2004, 20, 1–28 CrossRef CAS PubMed.
- H. Hotani and H. Miyamoto, Adv. Biophys., 1990, 26, 135–156 CrossRef CAS.
- D. K. Fygenson, J. F. Marko and A. Libchaber, Phys. Rev. Lett., 1997, 22, 4497–4500 CrossRef.
- B. Božič, S. Svetina and B. Žekš, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 1997, 55, 5834–5842 CrossRef.
- V. Heinrich, B. Božič, S. Svetina and B. Žekš, Biophys. J., 1999, 76, 2056–2071 CrossRef CAS.
- S. Svetina, B. Božič and B. Žekš, Eur. Biophys. J., 1998, 28, 74–77 CrossRef.
- G. H. W. Lim, G. Huber, Y. Torii, A. Hirata, J. Miller and S. Sazer, PLoS One, 2007, 9, e948 Search PubMed.
- M. Toya, M. Sato, U. Haselmann, K. Asakawa, D. Brunner, C. Antony and T. Toda, Nat. Cell Biol., 2007, 9, 646–653 CrossRef CAS PubMed.
- R. R. West, E. V. Vaisberg, P. Nurse and J. R. McIntosh, Mol. Biol. Cell, 1998, 9, 2839–2855 CrossRef CAS.
- A. Khodjakov, S. La Terra and F. Chang, Curr. Biol., 2004, 14, 1350–1360 CrossRef PubMed.
- H. Funabiki, I. Hagan, S. Uzawa and M. Yanagida, J. Cell Biol., 1993, 121, 961–976 CrossRef CAS.
- M. C. King, T. G. Drivas and G. Blobel, Cell, 2008, 134, 427–438 CrossRef CAS PubMed.
- I. M. Cheeseman, J. S. Chappie, E. M. Wilson-Kubalek and A. Desai, Cell, 2006, 127, 983–997 CrossRef CAS PubMed.
- H. Appelgren, B. Kniola and K. Ekwall, J. Cell Sci., 2003, 116, 4035–4042 CrossRef CAS PubMed.
- A. Nabetani, T. Koujin and C. Tsutsumi, et al. , Chromosoma, 2001, 5, 322–334 CrossRef PubMed.
- W. Helfrich, Z. Naturforsch., C: J. Biosci., 1973, 28, 693–703 CAS.
- Y. Gonzales, K. Meerbrey, J. Chong, Y. Torii, N. N. Padte and S. Sazer, J. Cell Sci., 2009, 122, 2464–2472 CrossRef PubMed.
- D. Zhang and S. Oliferenko, Curr. Opin. Cell Biol., 2010, 25, 142–148 CrossRef PubMed.
- S. Svetina and B. Žekš, Eur. Biophys. J., 1992, 21, 251–255 CrossRef CAS.
- M. G. Poirier, S. Eroglu and J. F. Marko, Mol. Biol. Cell, 2012, 13, 2170–2179 Search PubMed.
- G. Goshima, S. Saitoh and M. Yanagida, Genes Dev., 1999, 13, 1664–1677 CrossRef CAS.
- S. Saitoh, K. Takahashi, K. Nabeshima, Y. Yamashita, Y. Nakaseko, A. Hirata and M. Yanagida, J. Cell Biol., 1996, 134, 949–961 CrossRef CAS.
- V. Vitkova, D. Mitkova and G. Staneva, Colloids Surf., A, 2014, 460, 191–195 CrossRef CAS PubMed.
- S. Moreno, A. Klar and P. Nurse, Methods Enzymol., 1991, 194, 795–823 CAS.
- J. Z. Y. Chen, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2012, 85, 061910 CrossRef.
| This journal is © the Owner Societies 2015 |